Binding mode analysis of ABCA7 for the prediction of novel Alzheimer's disease therapeutics
- PMID: 34976306
- PMCID: PMC8666613
- DOI: 10.1016/j.csbj.2021.11.035
Binding mode analysis of ABCA7 for the prediction of novel Alzheimer's disease therapeutics
Abstract
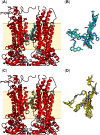
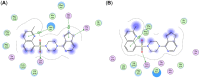
The adenosine-triphosphate-(ATP)-binding cassette (ABC) transporter ABCA7 is a genetic risk factor for Alzheimer's disease (AD). Defective ABCA7 promotes AD development and/or progression. Unfortunately, ABCA7 belongs to the group of 'under-studied' ABC transporters that cannot be addressed by small-molecules. However, such small-molecules would allow for the exploration of ABCA7 as pharmacological target for the development of new AD diagnostics and therapeutics. Pan-ABC transporter modulators inherit the potential to explore under-studied ABC transporters as novel pharmacological targets by potentially binding to the proposed 'multitarget binding site'. Using the recently reported cryogenic-electron microscopy (cryo-EM) structures of ABCA1 and ABCA4, a homology model of ABCA7 has been generated. A set of novel, diverse, and potent pan-ABC transporter inhibitors has been docked to this ABCA7 homology model for the discovery of the multitarget binding site. Subsequently, application of pharmacophore modelling identified the essential pharmacophore features of these compounds that may support the rational drug design of innovative diagnostics and therapeutics against AD.
Keywords: ABC transporter (ABCA1, ABCA4, ABCA7); ABC, ATP-binding cassette; AD, Alzheimer’s disease; APP, amyloid precursor protein; ATP, Adenosine-triphosphate; Alzheimer’s disease (AD); BBB, blood-brain barrier; BODIPY-cholesterol, 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-cholesterol; ECD, extracellular domain; EH, extracellular helix; GSH, reduced glutathione; HTS, high-throughput screening; IC, intracellular helix; MOE, Molecular Operating Environment; MSD, membrane spanning domain; Multitarget modulation (PANABC); NBD, nucleotide binding domain; NBD-cholesterol, 7-nitro-2-1,3-benzoxadiazol-4-yl-cholesterol; PDB, protein data bank; PET tracer (PETABC); PET, positron emission tomography; PLIF, protein ligand interaction; PSO, particle swarm optimization; Polypharmacology; R-domain/region, regulatory domain/region; RMSD, root mean square distance; Rational drug design and development; SNP, single-nucleotide polymorphism; TM, transmembrane helix; cryo-EM, cryogenic-electron microscopy.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Strategies to gain novel Alzheimer's disease diagnostics and therapeutics using modulators of ABCA transporters.Free Neuropathol. 2021;2:33. doi: 10.17879/freeneuropathology-2021-3528. Epub 2021 Dec 13. Free Neuropathol. 2021. PMID: 34977908 Free PMC article.
-
Physicochemistry shapes bioactivity landscape of pan-ABC transporter modulators: Anchor point for innovative Alzheimer's disease therapeutics.Int J Biol Macromol. 2022 Sep 30;217:775-791. doi: 10.1016/j.ijbiomac.2022.07.062. Epub 2022 Jul 13. Int J Biol Macromol. 2022. PMID: 35839956
-
ABCA7 Deficiency Accelerates Amyloid-β Generation and Alzheimer's Neuronal Pathology.J Neurosci. 2016 Mar 30;36(13):3848-59. doi: 10.1523/JNEUROSCI.3757-15.2016. J Neurosci. 2016. PMID: 27030769 Free PMC article.
-
ABCA7 links sterol metabolism to the host defense system: Molecular background for potential management measure of Alzheimer's disease.Gene. 2021 Feb 5;768:145316. doi: 10.1016/j.gene.2020.145316. Epub 2020 Nov 20. Gene. 2021. PMID: 33221536 Free PMC article. Review.
-
ABCA7 in Alzheimer's Disease.Mol Neurobiol. 2015;51(3):1008-16. doi: 10.1007/s12035-014-8759-9. Epub 2014 May 31. Mol Neurobiol. 2015. PMID: 24878767 Review.
Cited by
-
A curated binary pattern multitarget dataset of focused ATP-binding cassette transporter inhibitors.Sci Data. 2022 Jul 26;9(1):446. doi: 10.1038/s41597-022-01506-z. Sci Data. 2022. PMID: 35882865 Free PMC article.
-
Iron Overload in Brain: Transport Mismatches, Microbleeding Events, and How Nanochelating Therapies May Counteract Their Effects.Int J Mol Sci. 2024 Feb 16;25(4):2337. doi: 10.3390/ijms25042337. Int J Mol Sci. 2024. PMID: 38397013 Free PMC article. Review.
-
Indole Derivatives as New Structural Class of Potent and Antiproliferative Inhibitors of Monocarboxylate Transporter 1 (MCT1; SLC16A1).J Med Chem. 2023 Jan 12;66(1):657-676. doi: 10.1021/acs.jmedchem.2c01612. Epub 2022 Dec 30. J Med Chem. 2023. PMID: 36584238 Free PMC article.
-
The Necessity for Polypharmacological Research - An Editorial on 'Network Polypharmacology of ATP-binding Cassette (ABC) and Solute Carrier (SLC) Transporters'.Front Pharmacol. 2025 Mar 21;16:1561247. doi: 10.3389/fphar.2025.1561247. eCollection 2025. Front Pharmacol. 2025. PMID: 40201687 Free PMC article. No abstract available.
-
Medicinal polypharmacology-a scientific glossary of terminology and concepts.Front Pharmacol. 2024 Jul 18;15:1419110. doi: 10.3389/fphar.2024.1419110. eCollection 2024. Front Pharmacol. 2024. PMID: 39092220 Free PMC article. Review.
References
-
- Szakacs G., Abele R. An inventory of lysosomal ABC transporters. FEBS Lett. 2020;594:3965–3985. - PubMed
-
- Stefan K., Leck L.Y.W., Namasivayam V., Bascuñana P., Huang M.L.-H., Riss P.J., Pahnke J., Jansson P.J., Stefan S.M. Vesicular ATP-binding cassette transporters in human disease: relevant aspects of their organization for future drug development. Future Drug Discovery. 2020;2:FDD51.
-
- Stefan S.M., Jansson P.J., Kalinowski D.S., Anjum R., Dharmasivam M., Richardson D.R. The growing evidence for targeting P-glycoprotein in lysosomes to overcome resistance. Future Med Chem. 2020;12:473–477. - PubMed
LinkOut - more resources
Full Text Sources
