Yeast Crf1p: An activator in need is an activator indeed
- PMID: 34976315
- PMCID: PMC8688861
- DOI: 10.1016/j.csbj.2021.12.003
Yeast Crf1p: An activator in need is an activator indeed
Abstract
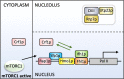
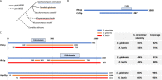
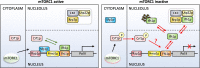
Ribosome biogenesis is an energetically costly process, and tight regulation is required for stoichiometric balance between components. This requires coordination of RNA polymerases I, II, and III. Lack of nutrients or the presence of stress leads to downregulation of ribosome biogenesis, a process for which mechanistic target of rapamycin complex I (mTORC1) is key. mTORC1 activity is communicated by means of specific transcription factors, and in yeast, which is a primary model system in which transcriptional coordination has been delineated, transcription factors involved in regulation of ribosomal protein genes include Fhl1p and its cofactors, Ifh1p and Crf1p. Ifh1p is an activator, whereas Crf1p has been implicated in maintaining the repressed state upon mTORC1 inhibition. Computational analyses of evolutionary relationships have indicated that Ifh1p and Crf1p descend from a common ancestor. Here, we discuss recent evidence, which suggests that Crf1p also functions as an activator. We propose a model that consolidates available experimental evidence, which posits that Crf1p functions as an alternate activator to prevent the stronger activator Ifh1p from re-binding gene promoters upon mTORC1 inhibition. The correlation between retention of Crf1p in related yeast strains and duplication of ribosomal protein genes suggests that this backup activation may be important to ensure gene expression when Ifh1p is limiting. With ribosome biogenesis as a hallmark of cell growth, failure to control assembly of ribosomal components leads to several human pathologies. A comprehensive understanding of mechanisms underlying this process is therefore of the essence.
Keywords: CK2, casein kinase 2; Crf1, corepressor with forkhead like; Crf1p; FHA, forkhead-associated; FHB, forkhead-binding; FKBP, FK506 binding protein; Fhl1, forkhead like; Fpr1, FK506-sensitive proline rotamase; Gene regulation; Hmo1, high mobility group; Ifh1, interacts with forkhead like; Ifh1p; RASTR, ribosome assembly stress response; RP, ribosomal protein; Rap1, repressor/activator protein; RiBi, ribosome biogenesis; Ribosomal protein; Ribosome biogenesis; Sfp1, split finger protein; WGD, whole genome duplication; mTORC1; mTORC1, mechanistic target of rapamycin complex 1.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Yeast Crf1p is an activator with different roles in regulation of target genes.Yeast. 2024 Jun;41(6):379-400. doi: 10.1002/yea.3939. Epub 2024 Apr 19. Yeast. 2024. PMID: 38639144
-
Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals.Biochem Soc Trans. 2021 Aug 27;49(4):1589-1599. doi: 10.1042/BST20201136. Biochem Soc Trans. 2021. PMID: 34240738 Free PMC article. Review.
-
Fpr1, a primary target of rapamycin, functions as a transcription factor for ribosomal protein genes cooperatively with Hmo1 in Saccharomyces cerevisiae.PLoS Genet. 2020 Jun 30;16(6):e1008865. doi: 10.1371/journal.pgen.1008865. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32603360 Free PMC article.
-
Physiological function of FKBP12, a primary target of rapamycin/FK506: a newly identified role in transcription of ribosomal protein genes in yeast.Curr Genet. 2021 Jun;67(3):383-388. doi: 10.1007/s00294-020-01142-3. Epub 2021 Jan 12. Curr Genet. 2021. PMID: 33438053 Review.
-
Role of CK2-dependent phosphorylation of Ifh1 and Crf1 in transcriptional regulation of ribosomal protein genes in Saccharomyces cerevisiae.Biochim Biophys Acta. 2016 Aug;1859(8):1004-13. doi: 10.1016/j.bbagrm.2016.06.003. Epub 2016 Jun 15. Biochim Biophys Acta. 2016. PMID: 27321754
Cited by
-
Obtaining new brewing yeasts using regional Chilean wine yeasts through an adaptive evolution program.Front Microbiol. 2025 Jun 16;16:1599904. doi: 10.3389/fmicb.2025.1599904. eCollection 2025. Front Microbiol. 2025. PMID: 40589584 Free PMC article.
-
Quiescence in Saccharomyces cerevisiae.Annu Rev Genet. 2022 Nov 30;56:253-278. doi: 10.1146/annurev-genet-080320-023632. Annu Rev Genet. 2022. PMID: 36449357 Free PMC article. Review.
-
The Central FacilitaTOR: Coordinating Transcription and Translation in Eukaryotes.Int J Mol Sci. 2025 Mar 21;26(7):2845. doi: 10.3390/ijms26072845. Int J Mol Sci. 2025. PMID: 40243440 Free PMC article. Review.
-
The Deletion of LeuRS Revealed Its Important Roles in Osmotic Stress Tolerance, Amino Acid and Sugar Metabolism, and the Reproduction Process of Aspergillus montevidensis.J Fungi (Basel). 2024 Jan 3;10(1):36. doi: 10.3390/jof10010036. J Fungi (Basel). 2024. PMID: 38248946 Free PMC article.
References
-
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. - PubMed
-
- Baßler J., Hurt E. Eukaryotic ribosome assembly. Annu Rev Biochem. 2019;88(1):281–306. - PubMed
-
- Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
