Classification of COVID-19 and Other Pathogenic Sequences: A Dinucleotide Frequency and Machine Learning Approach
- PMID: 34976561
- PMCID: PMC8675546
- DOI: 10.1109/ACCESS.2020.3031387
Classification of COVID-19 and Other Pathogenic Sequences: A Dinucleotide Frequency and Machine Learning Approach
Abstract
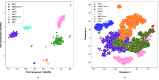
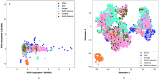
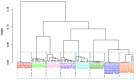
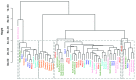
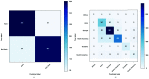
The world is grappling with the COVID-19 pandemic caused by the 2019 novel SARS-CoV-2. To better understand this novel virus and its relationship with other pathogens, new methods for analyzing the genome are required. In this study, intrinsic dinucleotide genomic signatures were analyzed for whole genome sequence data of eight pathogenic species, including SARS-CoV-2. The genome sequences were transformed into dinucleotide relative frequencies and classified using the extreme gradient boosting (XGBoost) model. The classification models were trained to a) distinguish between the sequences of all eight species and b) distinguish between sequences of SARS-CoV-2 that originate from different geographic regions. Our method attained 100% in all performance metrics and for all tasks in the eight-species classification problem. Moreover, the models achieved 67% balanced accuracy for the task of classifying the SARS-CoV-2 sequences into the six continental regions and achieved 86% balanced accuracy for the task of classifying SARS-CoV-2 samples as either originating from Asia or not. Analysis of the dinucleotide genomic profiles of the eight species revealed a similarity between the SARS-CoV-2 and MERS-CoV viral sequences. Further analysis of SARS-CoV-2 viral sequences from the six continents revealed that samples from Oceania had the highest frequency of TT dinucleotides as well as the lowest CG frequency compared to the other continents. The dinucleotide signatures of AC, AG,CA, CT, GA, GT, TC, and TG were well conserved across most genomes, while the frequencies of other dinucleotide signatures varied considerably. Altogether, the results from this study demonstrate the utility of dinucleotide relative frequencies for discriminating and identifying similar species.
Keywords: Alignment-free sequence analysis; COVID-19; XGBoost; dinucleotide frequencies; feature representations; genomic signatures; human pathogens; machine learning.
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 License. For more information, see https://creativecommons.org/licenses/by-nc-nd/4.0/.
Figures
Similar articles
-
Conservation vs. variation of dinucleotide frequencies across bacterial and archaeal genomes: evolutionary implications.Front Microbiol. 2013 Sep 6;4:269. doi: 10.3389/fmicb.2013.00269. eCollection 2013. Front Microbiol. 2013. PMID: 24046767 Free PMC article.
-
Conserved and varied dinucleotide sequences in the genomes of three Aspergillus species.Recent Adv DNA Gene Seq. 2014;8(1):10-4. doi: 10.2174/2352092208666141013231001. Recent Adv DNA Gene Seq. 2014. PMID: 25564023
-
Profiling SARS-CoV-2 mutation fingerprints that range from the viral pangenome to individual infection quasispecies.Genome Med. 2021 Apr 19;13(1):62. doi: 10.1186/s13073-021-00882-2. Genome Med. 2021. PMID: 33875001 Free PMC article.
-
A comparative genomics-based study of positive strand RNA viruses emphasizing on SARS-CoV-2 utilizing dinucleotide signature, codon usage and codon context analyses.Gene Rep. 2021 Jun;23:101055. doi: 10.1016/j.genrep.2021.101055. Epub 2021 Feb 17. Gene Rep. 2021. PMID: 33615042 Free PMC article.
-
Role of biological Data Mining and Machine Learning Techniques in Detecting and Diagnosing the Novel Coronavirus (COVID-19): A Systematic Review.J Med Syst. 2020 May 25;44(7):122. doi: 10.1007/s10916-020-01582-x. J Med Syst. 2020. PMID: 32451808 Free PMC article.
Cited by
-
Correlation-Based Analysis of COVID-19 Virus Genome Versus Other Fatal Virus Genomes.Arab J Sci Eng. 2021 Jun 24:1-13. doi: 10.1007/s13369-021-05811-4. Online ahead of print. Arab J Sci Eng. 2021. PMID: 34189012 Free PMC article.
-
Utilizing genomic signatures to gain insights into the dynamics of SARS-CoV-2 through Machine and Deep Learning techniques.BMC Bioinformatics. 2024 Mar 27;25(1):131. doi: 10.1186/s12859-024-05648-2. BMC Bioinformatics. 2024. PMID: 38539073 Free PMC article.
-
A Survey on Machine Learning and Internet of Medical Things-Based Approaches for Handling COVID-19: Meta-Analysis.Front Public Health. 2022 Jun 23;10:869238. doi: 10.3389/fpubh.2022.869238. eCollection 2022. Front Public Health. 2022. PMID: 35812486 Free PMC article.
-
PRCFX-DT: a new graph-based approach for feature selection and classification of genomic sequences.BMC Bioinformatics. 2025 Jun 17;26(1):159. doi: 10.1186/s12859-025-06183-4. BMC Bioinformatics. 2025. PMID: 40528202 Free PMC article.
-
A hybrid computational framework for intelligent inter-continent SARS-CoV-2 sub-strains characterization and prediction.Sci Rep. 2021 Jul 15;11(1):14558. doi: 10.1038/s41598-021-93757-w. Sci Rep. 2021. PMID: 34267263 Free PMC article.
References
-
- WHO. WHO coronavirus Disease (COVID-19) Dashboard [Online Dashboard]. Accessed: Jul. 23, 2020. [Online]. Available: https://covid19.who.int/
LinkOut - more resources
Full Text Sources
Miscellaneous
