Comprehensive analysis of N6-methyladenosine (m6A) modification during the degeneration of lumbar intervertebral disc in mice
- PMID: 34976732
- PMCID: PMC8685911
- DOI: 10.1016/j.jot.2021.10.008
Comprehensive analysis of N6-methyladenosine (m6A) modification during the degeneration of lumbar intervertebral disc in mice
Abstract
Objective: To study the N6-methyladenosine (m6A) modification pattern of nucleus pulposus (NP) tissue during intervertebral disc degeneration (IDD).
Methods: A standing mouse model was generated, and staining and imaging methods were used to evaluate the IDD model. Methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-seq) was used to analyze m6A methylation-associated transcripts in the NP, and real-time quantitative polymerase chain reaction (qRT-PCR) was used to detect the expression of methylation-related enzymes and conduct bio-informatics analysis.
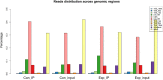
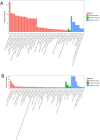
Results: The standing mouse model caused IDD. Continuous axial pressure changed the expression of related methylases in degenerated NP tissue. Relative to the control group, the expression levels of KIAA1429, METTL14, METTL3, METTL4, WTAP, DGCR8, EIF3A and YTHDC1 in the experimental group were higher, while those of FTO, ELAVL1, HNRNPC1 and SRSF2 were lower. We identified 985 differentially expressed genes through MeRIP-Seq, among which 363 genes were significantly up-regulated, and 622 genes were significantly down-regulated. In addition, among the 9648 genes counted, 1319 m6A peaks with significant differences in methylation were identified, among which 933 were significantly up-regulated, and 386 were significantly down-regulated. Genes and pathways that were enriched in IDD have been identified.
Conclusion: The results of this study elucidated the m6A methylation pattern of NP tissue in degenerated lumbar intervertebral disc of mice and provided new perspectives and clues for research on and the treatment of lumbar disc degeneration.
The translational potential of this article: As one of the important causes of low back and leg pain, intervertebral disc degeneration brings a huge economic burden to the society, family and medical system. Therefore, understanding the molecular and cellular mechanisms of intervertebral disc degeneration is of great significance for guiding clinical treatment. In this study, methylated RNA immunoprecipitation with next-generation sequencing on mice lumbar nucleus pulposus tissues found that differentially expressed genes and changes in the expression of related methylases, confirming that RNA methylation is involved in intervertebral disc degeneration. The process provides new vision and clues for future research on intervertebral disc degeneration.
Keywords: Intervertebral disc degeneration; Methylated RNA immunoprecipitation with next-generation sequencing; m6A methylation; nucleus pulposus.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Brodke D., Ritter S. Nonsurgical management of low back pain and lumbar disk degeneration. Instr Course Lect. 2005;54:279–286. - PubMed
-
- Bogduk N. Evidence-informed management of chronic low back pain with facet injections and radiofrequency neurotomy. Spine J : official journal of the North American Spine Society. 2008;8(1):56–64. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

