High ATF4 Expression Is Associated With Poor Prognosis, Amino Acid Metabolism, and Autophagy in Gastric Cancer
- PMID: 34976799
- PMCID: PMC8718699
- DOI: 10.3389/fonc.2021.740120
High ATF4 Expression Is Associated With Poor Prognosis, Amino Acid Metabolism, and Autophagy in Gastric Cancer
Abstract
Background: The role of activating transcription factor 4 (ATF4) underlying gastric cancer (GC) remains unclear. The purpose of this study was to investigate the expression levels and biological functions of ATF4 in GC.
Methods: Expression of ATF4 was detected by quantitative PCR (qPCR), Western blotting, and immunohistochemistry. Cox regression was used for survival analysis and the construction of the nomogram. Immunofluorescence was used to identify the intracellular localization of ATF4. Knockdown and overexpression of ATF4 in GC cells followed by wound healing and Transwell assays, EdU and Calcein-AM/propidium iodide (PI) staining, and cell cycle detection were performed to examine its function in vitro. Transmission electron microscopy was performed to assess the autophagy levels upon ATF4 silencing. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and gene set enrichment analysis (GSEA) were used to determine gene enrichment. SPSS 22.0 software, GraphPad Prism 7.0, and R version 3.6.1 were used for statistical analysis.
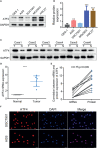
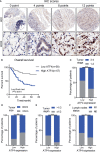
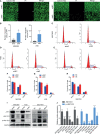
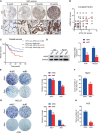
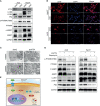
Results: ATF4 expression was upregulated in GC cells and tissues compared with corresponding normal tissues. Survival analysis suggested that a high ATF4 expression was strongly associated with worse overall survival (OS) of GC patients (p < 0.001). The nomogram and the receiver operating characteristic (ROC) curves demonstrated that ATF4 was a highly sensitive and specific prognostic marker of GC [C-index = 0.797, area under the ROC curve (AUC) of 3-year OS = 0.855, and AUC of 5-year OS = 0.863]. In addition, ATF4 knockdown inhibited the cell proliferation, migration, invasion, and cell cycle progression of GC cells in vitro, while overexpression of ATF4 exerted the opposite effects. Bioinformatics analysis showed that ATF4 could promote GC progression possibly by regulating asparagine (Asn) metabolism and autophagy pathways. Further experiments indicated that ATF4 expression was significantly positively correlated with ASNS expression. The inhibition of cell clone formation in Asn-deprived conditions was more significant in the shATF4 group. Finally, we found that ATF4 promoted autophagy through regulating the mTORC1 pathway in GC cells.
Conclusion: These findings suggested that ATF4 can significantly promote GC development and serve as an independent prognostic factor for GC.
Keywords: ATF4; activating transcription factor 4; autophagy; gastric cancer; metabolism; nomogram.
Copyright © 2021 Wang, Lu, Wang, Wu, Xu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Muro K, Van Cutsem E, Narita Y, Pentheroudakis G, Baba E, Li J, et al. . Pan-Asian Adapted ESMO Clinical Practice Guidelines for the Management of Patients With Metastatic Gastric Cancer: A JSMO-ESMO Initiative Endorsed by CSCO, KSMO, MOS, SSO and TOS. Ann Oncol (2019) 30(1):19–33. doi: 10.1093/annonc/mdy502 - DOI - PubMed
-
- Shitara K, Doi T, Dvorkin M, Mansoor W, Arkenau HT, Prokharau A, et al. . Trifluridine/tipiracil Versus Placebo in Patients With Heavily Pretreated Metastatic Gastric Cancer (TAGS): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol (2018) 19(11):1437–48. doi: 10.1016/S1470-2045(18)30739-3 - DOI - PubMed
-
- Fuchs CS, Doi T, Jang RW, Muro K, Satoh T, Machado M, et al. . Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol (2018) 4(5):e180013. doi: 10.1001/jamaoncol.2018.0013 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

