Capturing a Comprehensive Picture of Biological Events From Adverse Outcome Pathways in the Drug Exposome
- PMID: 34976924
- PMCID: PMC8718398
- DOI: 10.3389/fpubh.2021.763962
Capturing a Comprehensive Picture of Biological Events From Adverse Outcome Pathways in the Drug Exposome
Abstract
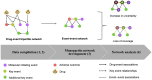
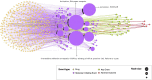
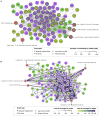
Background: The chemical part of the exposome, including drugs, may explain the increase of health effects with outcomes such as infertility, allergies, metabolic disorders, which cannot be only explained by the genetic changes. To better understand how drug exposure can impact human health, the concepts of adverse outcome pathways (AOPs) and AOP networks (AONs), which are representations of causally linked events at different biological levels leading to adverse health, could be used for drug safety assessment. Methods: To explore the action of drugs across multiple scales of the biological organization, we investigated the use of a network-based approach in the known AOP space. Considering the drugs and their associations to biological events, such as molecular initiating event and key event, a bipartite network was developed. This bipartite network was projected into a monopartite network capturing the event-event linkages. Nevertheless, such transformation of a bipartite network to a monopartite network had a huge risk of information loss. A way to solve this problem is to quantify the network reduction. We calculated two scoring systems, one measuring the uncertainty and a second one describing the loss of coverage on the developed event-event network to better investigate events from AOPs linked to drugs. Results: This AON analysis allowed us to identify biological events that are highly connected to drugs, such as events involving nuclear receptors (ER, AR, and PXR/SXR). Furthermore, we observed that the number of events involved in a linkage pattern with drugs is a key factor that influences information loss during monopartite network projection. Such scores have the potential to quantify the uncertainty of an event involved in an AON, and could be valuable for the weight of evidence assessment of AOPs. A case study related to infertility, more specifically to "decrease, male agenital distance" is presented. Conclusion: This study highlights that computational approaches based on network science may help to understand the complexity of drug health effects, with the aim to support drug safety assessment.
Keywords: AON; AOP; NAM; bipartite network; drug-AOP; network science.
Copyright © 2021 Wu, Bagdad, Taboureau and Audouze.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
An integrative data-centric approach to derivation and characterization of an adverse outcome pathway network for cadmium-induced toxicity.Sci Total Environ. 2024 Apr 10;920:170968. doi: 10.1016/j.scitotenv.2024.170968. Epub 2024 Feb 16. Sci Total Environ. 2024. PMID: 38367714
-
Investigation of a derived adverse outcome pathway (AOP) network for endocrine-mediated perturbations.Sci Total Environ. 2022 Jun 20;826:154112. doi: 10.1016/j.scitotenv.2022.154112. Epub 2022 Feb 24. Sci Total Environ. 2022. PMID: 35219661
-
Adverse outcome pathways and linkages to transcriptomic effects relevant to ionizing radiation injury.Int J Radiat Biol. 2022;98(12):1789-1801. doi: 10.1080/09553002.2022.2110313. Epub 2022 Aug 22. Int J Radiat Biol. 2022. PMID: 35939063
-
Building a Network of Adverse Outcome Pathways (AOPs) Incorporating the Tau-Driven AOP Toward Memory Loss (AOP429).J Alzheimers Dis Rep. 2022 Jun 7;6(1):271-296. doi: 10.3233/ADR-220015. eCollection 2022. J Alzheimers Dis Rep. 2022. PMID: 35891639 Free PMC article. Review.
-
Adverse outcome pathways: Application to enhance mechanistic understanding of neurotoxicity.Pharmacol Ther. 2017 Nov;179:84-95. doi: 10.1016/j.pharmthera.2017.05.006. Epub 2017 May 18. Pharmacol Ther. 2017. PMID: 28529068 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

