Using RNA-Seq to Investigate Immune-Metabolism Features in Immunocompromised Patients With Sepsis
- PMID: 34977060
- PMCID: PMC8718501
- DOI: 10.3389/fmed.2021.747263
Using RNA-Seq to Investigate Immune-Metabolism Features in Immunocompromised Patients With Sepsis
Abstract
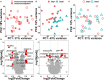
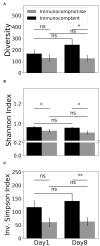
Objective: Sepsis is life threatening and leads to complex inflammation in patients with immunocompromised conditions, such as cancer, and receiving immunosuppressants for autoimmune diseases and organ transplant recipients. Increasing evidence has shown that RNA-Sequencing (RNA-Seq) can be used to define subendotype in patients with sepsis; therefore, we aim to use RNA-Seq to identify transcriptomic features among immunocompromised patients with sepsis. Methods: We enrolled patients who were admitted to medical intensive care units (ICUs) for sepsis at a tertiary referral centre in central Taiwan. Whole blood on day-1 and day-8 was obtained for RNA-Seq. We used Gene Set Enrichment Analysis (GSEA) to identify the enriched pathway of day-8/day-1 differentially expressed genes and MiXCR to determine the diversity of T cell repertoire. Results: A total of 18 immunocompromised subjects with sepsis and 18 sequential organ failure assessment (SOFA) score-matched immunocompetent control subjects were enrolled. The ventilator-day, ICU-stay, and hospital-day were similar between the two groups, whereas the hospital mortality was higher in immunocompromised patients than those in immunocompetent patients (50.0 vs. 5.6%, p < 0.01). We found that the top day-8/day-1 upregulated genes in the immunocompetent group were mainly innate immunity and inflammation relevant genes, namely, PRSS33, HDC, ALOX15, FCER1A, and OLR1, whereas a blunted day-8/day-1 dynamic transcriptome was found among immunocompromised patients with septic. Functional pathway analyses of day-8/day-1 differentially expressed genes identified the upregulated functional biogenesis and T cell-associated pathways in immunocompetent patients recovered from sepsis, whereas merely downregulated metabolism-associated pathways were found in immunocompromised patients with septic. Moreover, we used MiXCR to identify a higher diversity of T cell receptor (TCR) in immunocompetent patients both on day-1 and on day-8 than those in immunocompromised patients. Conclusions: Using RNA-Seq, we found compromised T cell function, altered metabolic signalling, and decreased T cell diversity among immunocompromised patients with septic, and more mechanistic studies are warranted to elucidate the underlying mechanism.
Keywords: RNA-Seq; immune; immunocompromised; metabolism; pathway analyses; sepsis.
Copyright © 2021 Cheng, Chen, Jiang, Hsiao, Wang, Wu, Ko and Chao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Whole transcriptome analysis to explore the impaired immunological features in critically ill elderly patients with sepsis.J Transl Med. 2023 Feb 23;21(1):141. doi: 10.1186/s12967-023-04002-z. J Transl Med. 2023. PMID: 36823620 Free PMC article.
-
Plasma protein C levels in immunocompromised septic patients are significantly lower than immunocompetent septic patients: a prospective cohort study.J Hematol Oncol. 2009 Oct 19;2:43. doi: 10.1186/1756-8722-2-43. J Hematol Oncol. 2009. PMID: 19840396 Free PMC article.
-
Identification of a transcriptome profile associated with improvement of organ function in septic shock patients after early supportive therapy.Crit Care. 2018 Nov 21;22(1):312. doi: 10.1186/s13054-018-2242-3. Crit Care. 2018. PMID: 30463588 Free PMC article.
-
[Clinical characteristics and prognosis of acute kidney injury in elderly patients with sepsis].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2019 Jul;31(7):837-841. doi: 10.3760/cma.j.issn.2095-4352.2019.07.008. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2019. PMID: 31441406 Chinese.
-
[Lactic acid, lactate clearance and procalcitonin in assessing the severity and predicting prognosis in sepsis].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2020 Apr;32(4):449-453. doi: 10.3760/cma.j.cn121430-20200129-00086. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2020. PMID: 32527351 Chinese.
Cited by
-
Identification of hub genes for adult patients with sepsis via RNA sequencing.Sci Rep. 2022 Mar 24;12(1):5128. doi: 10.1038/s41598-022-09175-z. Sci Rep. 2022. PMID: 35332254 Free PMC article.
-
The Association Between Absolute Lymphocyte Count and Long-Term Mortality in Critically Ill Medical Patients: Propensity Score-Based Analyses.Int J Gen Med. 2023 Aug 22;16:3665-3675. doi: 10.2147/IJGM.S424724. eCollection 2023. Int J Gen Med. 2023. PMID: 37637708 Free PMC article.
-
Bioinformatics Analysis of Gene Expression Profiles for Diagnosing Sepsis and Risk Prediction in Patients with Sepsis.Int J Mol Sci. 2023 May 27;24(11):9362. doi: 10.3390/ijms24119362. Int J Mol Sci. 2023. PMID: 37298316 Free PMC article.
-
Integration of Network Pharmacology, Transcriptomics, and Metabolomics Strategies to Uncover the Mechanism of Chaihuang Qingfu Pill in Treating Sepsis-Induced Liver Injury.Drug Des Devel Ther. 2025 Jun 2;19:4665-4688. doi: 10.2147/DDDT.S521626. eCollection 2025. Drug Des Devel Ther. 2025. PMID: 40486124 Free PMC article.
-
Whole transcriptome analysis to explore the impaired immunological features in critically ill elderly patients with sepsis.J Transl Med. 2023 Feb 23;21(1):141. doi: 10.1186/s12967-023-04002-z. J Transl Med. 2023. PMID: 36823620 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

