An Introduction to Machine Learning Approaches for Biomedical Research
- PMID: 34977072
- PMCID: PMC8716730
- DOI: 10.3389/fmed.2021.771607
An Introduction to Machine Learning Approaches for Biomedical Research
Abstract
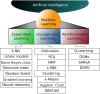
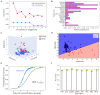
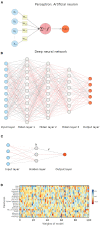
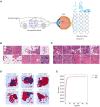
Machine learning (ML) approaches are a collection of algorithms that attempt to extract patterns from data and to associate such patterns with discrete classes of samples in the data-e.g., given a series of features describing persons, a ML model predicts whether a person is diseased or healthy, or given features of animals, it predicts weather an animal is treated or control, or whether molecules have the potential to interact or not, etc. ML approaches can also find such patterns in an agnostic manner, i.e., without having information about the classes. Respectively, those methods are referred to as supervised and unsupervised ML. A third type of ML is reinforcement learning, which attempts to find a sequence of actions that contribute to achieving a specific goal. All of these methods are becoming increasingly popular in biomedical research in quite diverse areas including drug design, stratification of patients, medical images analysis, molecular interactions, prediction of therapy outcomes and many more. We describe several supervised and unsupervised ML techniques, and illustrate a series of prototypical examples using state-of-the-art computational approaches. Given the complexity of reinforcement learning, it is not discussed in detail here, instead, interested readers are referred to excellent reviews on that topic. We focus on concepts rather than procedures, as our goal is to attract the attention of researchers in biomedicine toward the plethora of powerful ML methods and their potential to leverage basic and applied research programs.
Keywords: biomedical research; machine learning; reinforcement learning; supervised learning; unsupervised learning.
Copyright © 2021 Jovel and Greiner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition. New York, NY: Springer Science & Business Media; (2009).
-
- Müller AC, Guido S. Introduction to Machine Learning with Python: A Guide for Data Scientists. Sebastopol, CA: O'Reilly Media, Inc. (2016).
-
- Ayodele TO. Types of machine learning algorithms. New Adv Mach Learn. (2010) 3:19–48. 10.5772/9385 - DOI
-
- Berry MW, Mohamed A, Yap BW. Supervised and Unsupervised Learning for Data Science. Cham: Springer Nature; (2019). 10.1007/978-3-030-22475-2 - DOI
LinkOut - more resources
Full Text Sources

