Mendelian Randomization Integrating GWAS, eQTL, and mQTL Data Identified Genes Pleiotropically Associated With Atrial Fibrillation
- PMID: 34977172
- PMCID: PMC8719596
- DOI: 10.3389/fcvm.2021.745757
Mendelian Randomization Integrating GWAS, eQTL, and mQTL Data Identified Genes Pleiotropically Associated With Atrial Fibrillation
Abstract
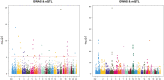
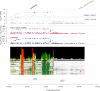
Background: Atrial fibrillation (AF) is the most common arrhythmia. Genome-wide association studies (GWAS) have identified more than 100 loci associated with AF, but the underlying biological interpretation remains largely unknown. The goal of this study is to identify gene expression and DNA methylation (DNAm) that are pleiotropically or potentially causally associated with AF, and to integrate results from transcriptome and methylome. Methods: We used the summary data-based Mendelian randomization (SMR) to integrate GWAS with expression quantitative trait loci (eQTL) studies and methylation quantitative trait loci (mQTL) studies. The HEIDI (heterogeneity in dependent instruments) test was introduced to test against the null hypothesis that there is a single causal variant underlying the association. Results: We prioritized 22 genes by eQTL analysis and 50 genes by mQTL analysis that passed the SMR & HEIDI test. Among them, 6 genes were overlapped. By incorporating consistent SMR associations between DNAm and AF, between gene expression and AF, and between DNAm and gene expression, we identified several mediation models at which a genetic variant exerted an effect on AF by altering the DNAm level, which regulated the expression level of a functional gene. One example was the genetic variant-cg18693985-CPEB4-AF axis. Conclusion: In conclusion, our integrative analysis identified multiple genes and DNAm sites that had potentially causal effects on AF. We also pinpointed plausible mechanisms in which the effect of a genetic variant on AF was mediated by genetic regulation of transcription through DNAm. Further experimental validation is necessary to translate the identified genes and possible mechanisms into clinical practice.
Keywords: GWAS; Mendelian randomization; atrial fibrillation; eQTL; mQTL; multi-omics.
Copyright © 2021 Liu, Li, Ma, Huang, Ouyang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Hindricks G, Potpara T, Dagres N, Arbelo E, Bax JJ, Blomström-Lundqvist C, et al. . 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association of Cardio-Thoracic Surgery (EACTS). Eur Heart J. (2020) 42:373–498. 10.1093/eurheartj/ehaa612 - DOI - PubMed
LinkOut - more resources
Full Text Sources

