β-catenin links cell seeding density to global gene expression during mouse embryonic stem cell differentiation
- PMID: 34977504
- PMCID: PMC8689156
- DOI: 10.1016/j.isci.2021.103541
β-catenin links cell seeding density to global gene expression during mouse embryonic stem cell differentiation
Abstract
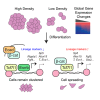
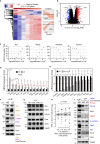
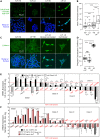
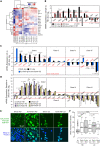
Although cell density is known to affect numerous biological processes including gene expression and cell fate specification, mechanistic understanding of what factors link cell density to global gene regulation is lacking. Here, we reveal that the expression of thousands of genes in mouse embryonic stem cells (mESCs) is affected by cell seeding density and that low cell density enhances the efficiency of differentiation. Mechanistically, β-catenin is localized primarily to adherens junctions during both self-renewal and differentiation at high density. However, when mESCs differentiate at low density, β-catenin translocates to the nucleus and associates with Tcf7l1, inducing co-occupied lineage markers. Meanwhile, Esrrb sustains the expression of pluripotency-associated genes while repressing lineage markers at high density, and its association with DNA decreases at low density. Our results provide new insights into the previously neglected but pervasive phenomenon of density-dependent gene regulation.
Keywords: Cell biology; Stem cells research; Transcriptomics.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Ectopic γ-catenin expression partially mimics the effects of stabilized β-catenin on embryonic stem cell differentiation.PLoS One. 2013 May 27;8(5):e65320. doi: 10.1371/journal.pone.0065320. Print 2013. PLoS One. 2013. PMID: 23724138 Free PMC article.
-
CHIR99021 enhances Klf4 Expression through β-Catenin Signaling and miR-7a Regulation in J1 Mouse Embryonic Stem Cells.PLoS One. 2016 Mar 3;11(3):e0150936. doi: 10.1371/journal.pone.0150936. eCollection 2016. PLoS One. 2016. PMID: 26938105 Free PMC article.
-
Disrupting Interactions Between β-Catenin and Activating TCFs Reconstitutes Ground State Pluripotency in Mouse Embryonic Stem Cells.Stem Cells. 2017 Aug;35(8):1924-1933. doi: 10.1002/stem.2647. Epub 2017 Jun 15. Stem Cells. 2017. PMID: 28577307
-
Inhibition of β-catenin-TCF1 interaction delays differentiation of mouse embryonic stem cells.J Cell Biol. 2015 Oct 12;211(1):39-51. doi: 10.1083/jcb.201503017. J Cell Biol. 2015. PMID: 26459597 Free PMC article.
-
β-Catenin in pluripotency: adhering to self-renewal or Wnting to differentiate?Int Rev Cell Mol Biol. 2014;312:53-78. doi: 10.1016/B978-0-12-800178-3.00002-6. Int Rev Cell Mol Biol. 2014. PMID: 25262238 Review.
Cited by
-
MicroRNA-329-3p inhibits the Wnt/β-catenin pathway and proliferation of osteosarcoma cells by targeting transcription factor 7-like 1.Oncol Res. 2024 Feb 6;32(3):463-476. doi: 10.32604/or.2023.044085. eCollection 2024. Oncol Res. 2024. PMID: 38370338 Free PMC article.
-
Role of the cytoskeleton in cellular reprogramming: effects of biophysical and biochemical factors.Front Mol Biosci. 2025 Mar 7;12:1538806. doi: 10.3389/fmolb.2025.1538806. eCollection 2025. Front Mol Biosci. 2025. PMID: 40123979 Free PMC article. Review.
-
Control of spatio-temporal patterning via cell growth in a multicellular synthetic gene circuit.Nat Commun. 2024 Nov 19;15(1):9867. doi: 10.1038/s41467-024-53078-8. Nat Commun. 2024. PMID: 39562554 Free PMC article.
-
Single Cell Transcriptomics-Informed Induced Pluripotent Stem Cells Differentiation to Tenogenic Lineage.bioRxiv [Preprint]. 2024 Jul 10:2023.04.10.536240. doi: 10.1101/2023.04.10.536240. bioRxiv. 2024. PMID: 37090543 Free PMC article. Preprint.
-
MDM2 inhibitors, nutlin-3a and navtemadelin, retain efficacy in human and mouse cancer cells cultured in hypoxia.Sci Rep. 2023 Mar 20;13(1):4583. doi: 10.1038/s41598-023-31484-0. Sci Rep. 2023. PMID: 36941277 Free PMC article.
References
-
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

