Insights into phosphatase-activated chemical defense in a marine sponge holobiont
- PMID: 34977575
- PMCID: PMC8637855
- DOI: 10.1039/d1cb00163a
Insights into phosphatase-activated chemical defense in a marine sponge holobiont
Abstract
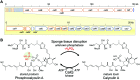
Marine sponges often contain potent cytotoxic compounds, which in turn evokes the principle question of how marine sponges avoid self-toxicity. In a marine sponge Discodermia calyx, the highly toxic calyculin A is detoxified by the phosphorylation, which is catalyzed by the phosphotransferase CalQ of a producer symbiont, "Candidatus Entotheonella" sp. Here we show the activating mechanism to dephosphorylate the stored phosphocalyculin A protoxin. The phosphatase specific to phosphocalyculin A is CalL, which is also encoded in the calyculin biosynthetic gene cluster. CalL represents a new clade and unprecedently coordinates the heteronuclear metals Cu and Zn. CalL is localized in the periplasmic space of the sponge symbiont, where it is ready for the on-demand production of calyculin A in response to sponge tissue disruption.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont.Nat Chem Biol. 2014 Aug;10(8):648-55. doi: 10.1038/nchembio.1573. Epub 2014 Jun 29. Nat Chem Biol. 2014. PMID: 24974231
-
Phosphocalyculin C as a pyrophosphate protoxin of calyculin C in the marine sponge Discodermia calyx.Bioorg Med Chem Lett. 2014 Nov 15;24(22):5150-3. doi: 10.1016/j.bmcl.2014.10.002. Epub 2014 Oct 13. Bioorg Med Chem Lett. 2014. PMID: 25442302
-
Biosynthesis of Bioactive Natural Products Derived from Theonellidae Family Marine Sponges.Chem Pharm Bull (Tokyo). 2023;71(1):1-8. doi: 10.1248/cpb.c22-00715. Chem Pharm Bull (Tokyo). 2023. PMID: 36596505 Review.
-
Calyculin: Nature's way of making the sponge-derived cytotoxin.Nat Prod Rep. 2016 Jun 2;33(6):751-60. doi: 10.1039/c5np00123d. Nat Prod Rep. 2016. PMID: 26923942 Review.
-
Biosynthetic Insights of Calyculin- and Misakinolide-Type Compounds in "Candidatus Entotheonella sp.".Methods Enzymol. 2018;604:287-330. doi: 10.1016/bs.mie.2018.02.017. Epub 2018 May 7. Methods Enzymol. 2018. PMID: 29779656
Cited by
-
Jaspamide/Jasplakinolide Is Synthesized by Jaspinella (Tectomicrobia) Bacteria in Sponges.J Nat Prod. 2025 Jun 27;88(6):1471-1480. doi: 10.1021/acs.jnatprod.5c00433. Epub 2025 Jun 4. J Nat Prod. 2025. PMID: 40468714
-
Impact of Marine Chemical Ecology Research on the Discovery and Development of New Pharmaceuticals.Mar Drugs. 2023 Mar 9;21(3):174. doi: 10.3390/md21030174. Mar Drugs. 2023. PMID: 36976223 Free PMC article. Review.
References
-
- Wright J. T. Benkendorff K. Davis A. R. J. Exp. Mar. Biol. Ecol. 1997;213:199–213. doi: 10.1016/S0022-0981(96)02768-2. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

