MATISSE: An analysis protocol for combining imaging mass cytometry with fluorescence microscopy to generate single-cell data
- PMID: 34977680
- PMCID: PMC8689354
- DOI: 10.1016/j.xpro.2021.101034
MATISSE: An analysis protocol for combining imaging mass cytometry with fluorescence microscopy to generate single-cell data
Abstract
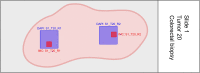
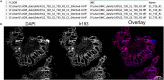
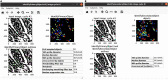
Exploring tissue heterogeneity on a single-cell level by imaging mass cytometry (IMC) remains challenging because of its limiting resolution. We previously demonstrated that combining higher resolution fluorescence with IMC data in the analysis pipeline resulted in high-quality single-cell segmentation. Here, we provide a step-by-step workflow of this MATISSE pipeline, including instructions regarding the staining procedure, and the analysis route to generate single-cell data. For complete details on the use and execution of this protocol, please refer to Baars et al., 2021.
Keywords: Antibody; Bioinformatics; Biotechnology and bioengineering; Cell Biology; Flow Cytometry/Mass Cytometry; Microscopy; Single Cell.
© 2021 The Author(s).
Conflict of interest statement
Y.V. receives funding through a Public-private partnership grant (TKI-Health Holland) with TigaTx B.V.
Figures











References
-
- Brown M., Szeliski R., Winder S. IEEE; 2005. Multi-image matching using multi-scale oriented Patches; pp. 510–517. (2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05)). - DOI
-
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

