Identification of Lipid Heterogeneity and Diversity in the Developing Human Brain
- PMID: 34977897
- PMCID: PMC8717369
- DOI: 10.1021/jacsau.1c00393
Identification of Lipid Heterogeneity and Diversity in the Developing Human Brain
Abstract
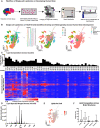
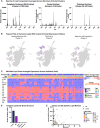
The lipidome is currently understudied but fundamental to life. Within the brain, little is known about cell-type lipid heterogeneity, and even less is known about cell-to-cell lipid diversity because it is difficult to study the lipids within individual cells. Here, we used single-cell mass spectrometry-based protocols to profile the lipidomes of 154 910 single cells across ten individuals consisting of five developmental ages and five brain regions, resulting in a unique lipid atlas available via a web browser of the developing human brain. From these data, we identify differentially expressed lipids across brain structures, cortical areas, and developmental ages. We inferred lipid profiles of several major cell types from this data set and additionally detected putative cell-type specific lipids. This data set will enable further interrogation of the developing human brain lipidome.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Marsh D.; Horvath L. I.; Swamy M. J.; Mantripragada S.; Kleinschmidt J. H. Interaction of membrane-spanning proteins with peripheral and lipid-anchored membrane proteins: perspectives from protein-lipid interactions (Review). Mol. Membr. Biol. 2002, 19 (4), 247–55. 10.1080/09687680210162419. - DOI - PubMed
