Accurate Machine Learning Prediction of Protein Circular Dichroism Spectra with Embedded Density Descriptors
- PMID: 34977905
- PMCID: PMC8715543
- DOI: 10.1021/jacsau.1c00449
Accurate Machine Learning Prediction of Protein Circular Dichroism Spectra with Embedded Density Descriptors
Abstract
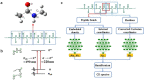
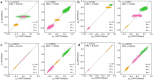
A data-driven approach to simulate circular dichroism (CD) spectra is appealing for fast protein secondary structure determination, yet the challenge of predicting electric and magnetic transition dipole moments poses a substantial barrier for the goal. To address this problem, we designed a new machine learning (ML) protocol in which ordinary pure geometry-based descriptors are replaced with alternative embedded density descriptors and electric and magnetic transition dipole moments are successfully predicted with an accuracy comparable to first-principle calculation. The ML model is able to not only simulate protein CD spectra nearly 4 orders of magnitude faster than conventional first-principle simulation but also obtain CD spectra in good agreement with experiments. Finally, we predicted a series of CD spectra of the Trp-cage protein associated with continuous changes of protein configuration along its folding path, showing the potential of our ML model for supporting real-time CD spectroscopy study of protein dynamics.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
LinkOut - more resources
Full Text Sources
