pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar
- PMID: 34977949
- PMCID: PMC8825253
- DOI: 10.1093/plphys/kiab546
pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar
Abstract
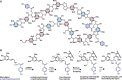
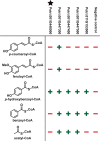
Poplar (Populus) lignin is naturally acylated with p-hydroxybenzoate ester moieties. However, the enzyme(s) involved in the biosynthesis of the monolignol-p-hydroxybenzoates have remained largely unknown. Here, we performed an in vitro screen of the Populus trichocarpa BAHD acyltransferase superfamily (116 genes) using a wheatgerm cell-free translation system and found five enzymes capable of producing monolignol-p-hydroxybenzoates. We then compared the transcript abundance of the five corresponding genes with p-hydroxybenzoate concentrations using naturally occurring unrelated genotypes of P. trichocarpa and revealed a positive correlation between the expression of p-hydroxybenzoyl-CoA monolig-nol transferase (pHBMT1, Potri.001G448000) and p-hydroxybenzoate levels. To test whether pHBMT1 is responsible for the biosynthesis of monolignol-p-hydroxybenzoates, we overexpressed pHBMT1 in hybrid poplar (Populus alba × P. grandidentata) (35S::pHBMT1 and C4H::pHBMT1). Using three complementary analytical methods, we showed that there was an increase in soluble monolignol-p-hydroxybenzoates and cell-wall-bound monolignol-p-hydroxybenzoates in the poplar transgenics. As these pendent groups are ester-linked, saponification releases p-hydroxybenzoate, a precursor to parabens that are used in pharmaceuticals and cosmetics. This identified gene could therefore be used to engineer lignocellulosic biomass with increased value for emerging biorefinery strategies.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures


References
-
- Bai Z, Phuan WC, Ding J, Heng TH, Luo J, Zhu Y (2016) Production of terephthalic acid from lignin-based phenolic acids by a cascade fixed-bed process. ACS Catal 6:6141–6145
-
- Beuerle T, Pichersky E (2002) Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem 302:305–312 - PubMed
-
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546 - PubMed
-
- Cesarino I (2019) Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr Opin Biotechnol 56:209–214 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

