Sustained, complete response to pexidartinib in a patient with CSF1R-mutated Erdheim-Chester disease
- PMID: 34978715
- PMCID: PMC9536810
- DOI: 10.1002/ajh.26441
Sustained, complete response to pexidartinib in a patient with CSF1R-mutated Erdheim-Chester disease
Abstract
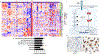
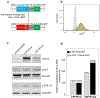
Erdheim-Chester disease (ECD) is a histiocytic neoplasm that predominantly harbors mitogen-activated protein kinase (MAPK) pathway variants. MAPK inhibitors typically are effective treatments, but mutations outside the MAPK pathway, such as CSF1R variants, may cause refractory ECD. We describe a patient with a novel somatic mutation in CSF1R (CSF1RR549_E554delinsQ ) that resulted in refractory ECD affecting the central nervous system. Cell model studies, RNA sequencing analysis, and in silico protein modeling suggested that she had a gain-of-function mutation occurring in a region critical for autoinhibition. The patient was treated with pexidartinib, a CSF1R inhibitor, and has had a complete clinical and metabolic response lasting more than 1.5 years to date. To our knowledge, this is the first report to describe successful treatment of a patient with ECD by using an agent that specifically targets CSF1R. This case also highlights the critical role of individualized molecular profiling to identify novel therapeutic targets in ECD.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
Conflicts of Interest
Authors have no conflicts of interest to disclose regarding this work.
Figures
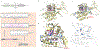

References
-
- Goyal G, Heaney ML, Collin M, et al. Erdheim-Chester disease: consensus recommendations for evaluation, diagnosis, and treatment in the molecular era. Blood. 2020;135(22):1929–1945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
