Investigation of Decitabine Effects on HDAC3 and HDAC7 mRNA Expression in NALM-6 and HL-60 Cancer Cell Lines
- PMID: 34981019
- PMCID: PMC8718785
- DOI: 10.52547/rbmb.10.3.420
Investigation of Decitabine Effects on HDAC3 and HDAC7 mRNA Expression in NALM-6 and HL-60 Cancer Cell Lines
Abstract
Background: Decitabine is a potent anticancer hypomethylating agent and changes the gene expression through the gene's promoter demethylation and also independently from DNA demethylation. So, the present study was designed to distinguish whether Decitabine, in addition to inhibitory effects on DNA methyltransferase, can change HDAC3 and HDAC7 mRNA expression in NALM-6 and HL-60 cancer cell lines.
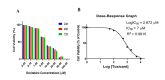
Methods: HL-60, NALM-6, and normal cells were cultured, and the Decitabine treatment dose was obtained (1 µM) through the MTT assay. Finally, HDAC3 and HDAC7 mRNA expression were measured by Real-Time PCR in HL-60 and NALM-6 cancerous cells before and after treatment. Furthermore, HDAC3 and HDAC7 mRNA expression in untreated HL-60 and NALM-6 cancerous cells were compared to normal cells.
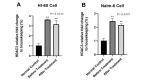
Results: Our results revealed that the expression of HDAC3 and HDAC7 in HL-60 and NALM-6 cells increases as compared to normal cells. After treatment of HL-60 and NALM-6 cells with Decitabine, HDAC3, and HDAC7 mRNA expression were decreased significantly.
Conclusion: Our data confirmed that the effects of Decitabine are not limited to direct hypomethylation of DNMTs, but it can indirectly affect other epigenetic factors, such as HDACs activity, through converging pathways.
Keywords: Decitabine; HDAC3; HDAC7; HL-60; NALM-6.
Figures


References
-
- Abazari O, Shafaei Z, Divsalar A, Eslami-Moghadam M, Ghalandari B, Saboury AA. Probing the biological evaluations of a new designed Pt (II) complex using spectroscopic and theoretical approaches: Human hemoglobin as a target. . J Biomol Struct Dyn. . 2016;34(5):1123, 31. - PubMed
LinkOut - more resources
Full Text Sources
