This is a preprint.
The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters
- PMID: 34981044
- PMCID: PMC8722607
- DOI: 10.21203/rs.3.rs-1211792/v1
The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters
Update in
-
SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters.Nature. 2022 Mar;603(7902):687-692. doi: 10.1038/s41586-022-04441-6. Epub 2022 Jan 21. Nature. 2022. PMID: 35062015 Free PMC article.
Abstract
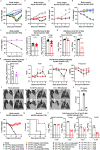
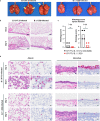
Despite the development and deployment of antibody and vaccine countermeasures, rapidly-spreading SARS-CoV-2 variants with mutations at key antigenic sites in the spike protein jeopardize their efficacy. The recent emergence of B.1.1.529, the Omicron variant1,2, which has more than 30 mutations in the spike protein, has raised concerns for escape from protection by vaccines and therapeutic antibodies. A key test for potential countermeasures against B.1.1.529 is their activity in pre-clinical rodent models of respiratory tract disease. Here, using the collaborative network of the SARS-CoV-2 Assessment of Viral Evolution (SAVE) program of the National Institute of Allergy and Infectious Diseases (NIAID), we evaluated the ability of multiple B.1.1.529 Omicron isolates to cause infection and disease in immunocompetent and human ACE2 (hACE2) expressing mice and hamsters. Despite modeling and binding data suggesting that B.1.1.529 spike can bind more avidly to murine ACE2, we observed attenuation of infection in 129, C57BL/6, and BALB/c mice as compared with previous SARS-CoV-2 variants, with limited weight loss and lower viral burden in the upper and lower respiratory tracts. Although K18-hACE2 transgenic mice sustained infection in the lungs, these animals did not lose weight. In wild-type and hACE2 transgenic hamsters, lung infection, clinical disease, and pathology with B.1.1.529 also were milder compared to historical isolates or other SARS-CoV-2 variants of concern. Overall, experiments from multiple independent laboratories of the SAVE/NIAID network with several different B.1.1.529 isolates demonstrate attenuated lung disease in rodents, which parallels preliminary human clinical data.
Figures

References
Publication types
Grants and funding
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- R01 AI157155/AI/NIAID NIH HHS/United States
- 75N93021C00017/AI/NIAID NIH HHS/United States
- U01 AI151810/AI/NIAID NIH HHS/United States
- 75N93021C00016/AI/NIAID NIH HHS/United States
- R01 AI129269/AI/NIAID NIH HHS/United States
- P01 AI060699/AI/NIAID NIH HHS/United States
- HHSN272201400004C/AI/NIAID NIH HHS/United States
- 75N93021C00014/AI/NIAID NIH HHS/United States
- R56 AI147623/AI/NIAID NIH HHS/United States
- 75N93019C00051/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- HHSN272201700041I/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

