This is a preprint.
Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses
- PMID: 34981049
- PMCID: PMC8722586
- DOI: 10.1101/2021.12.03.471045
Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses
Update in
-
SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.Cell. 2022 Feb 3;185(3):467-484.e15. doi: 10.1016/j.cell.2021.12.046. Epub 2022 Jan 4. Cell. 2022. PMID: 35081335 Free PMC article.
Abstract
On the 24 th November 2021 the sequence of a new SARS CoV-2 viral isolate spreading rapidly in Southern Africa was announced, containing far more mutations in Spike (S) than previously reported variants. Neutralization titres of Omicron by sera from vaccinees and convalescent subjects infected with early pandemic as well as Alpha, Beta, Gamma, Delta are substantially reduced or fail to neutralize. Titres against Omicron are boosted by third vaccine doses and are high in cases both vaccinated and infected by Delta. Mutations in Omicron knock out or substantially reduce neutralization by most of a large panel of potent monoclonal antibodies and antibodies under commercial development. Omicron S has structural changes from earlier viruses, combining mutations conferring tight binding to ACE2 to unleash evolution driven by immune escape, leading to a large number of mutations in the ACE2 binding site which rebalance receptor affinity to that of early pandemic viruses.
Conflict of interest statement
Competing Financial Interests
G.R.S sits on the GSK Vaccines Scientific Advisory Board and is a founder member of RQ Biotechnology. J.Z. and G.S. declare the Israel patent application no. 23/09/2020 – 277546 and U.S.A patent application no. 16/12/2020 – 63/125,984, entitled Methods and compositions for treating coronaviral infections. Oxford University holds intellectual property related to the Oxford-AstraZeneca vaccine. AJP is Chair of UK Dept. Health and Social Care’s (DHSC) Joint Committee on Vaccination & Immunisation (JCVI) but does not participate in the JCVI COVID19 committee, and is a member of the WHO’s SAGE. The views expressed in this article do not necessarily represent the views of DHSC, JCVI, or WHO. The University of Oxford has entered into a partnership with AstraZeneca on coronavirus vaccine development. The University of Oxford has protected intellectual property disclosed in this publication. S.C.G. is co-founder of Vaccitech (collaborators in the early development of this vaccine candidate) and is named as an inventor on a patent covering use of ChAdOx1-vectored vaccines and a patent application covering this SARS-CoV-2 vaccine (PCT/GB2012/000467). T.L. is named as an inventor on a patent application covering this SARS-CoV-2 vaccine and was a consultant to Vaccitech for an unrelated project during the conduct of the study. S.J.D. is a Scientific Advisor to the Scottish Parliament on COVID-19.
Figures

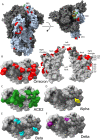

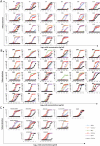

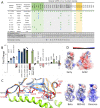

References
-
- Acharya R., Fry E., Stuart D., Fox G., Rowlands D., and Brown F. (1989). The three-dimensional structure of foot-and-mouth disease virus at 2.9 A resolution. Nature 337, 709–716. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
