This is a preprint.
SARS-CoV-2 spike conformation determines plasma neutralizing activity
- PMID: 34981060
- PMCID: PMC8722597
- DOI: 10.1101/2021.12.19.473391
SARS-CoV-2 spike conformation determines plasma neutralizing activity
Update in
-
SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines.Sci Immunol. 2022 Dec 23;7(78):eadf1421. doi: 10.1126/sciimmunol.adf1421. Epub 2022 Dec 23. Sci Immunol. 2022. PMID: 36356052 Free PMC article.
Abstract
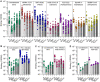
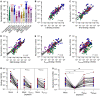
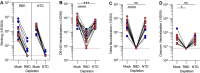
Numerous safe and effective COVID-19 vaccines have been developed that utilize various delivery technologies and engineering strategies. The influence of the SARS-CoV-2 spike (S) glycoprotein conformation on antibody responses induced by vaccination or infection in humans remains unknown. To address this question, we compared plasma antibodies elicited by six globally-distributed vaccines or infection and observed markedly higher binding titers for vaccines encoding a prefusion-stabilized S relative to other groups. Prefusion S binding titers positively correlated with plasma neutralizing activity, indicating that physical stabilization of the prefusion conformation enhances protection against SARS-CoV-2. We show that almost all plasma neutralizing activity is directed to prefusion S, in particular the S 1 subunit, and that variant cross-neutralization is mediated solely by RBD-specific antibodies. Our data provide a quantitative framework for guiding future S engineering efforts to develop vaccines with higher resilience to the emergence of variants and longer durability than current technologies.
Figures
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181, 271–280.e8 (2020). - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
