SNP characteristics and validation success in genome wide association studies
- PMID: 34981173
- PMCID: PMC8855685
- DOI: 10.1007/s00439-021-02407-8
SNP characteristics and validation success in genome wide association studies
Abstract
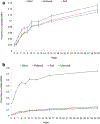
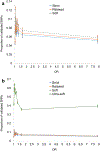
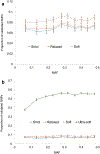
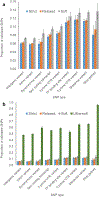
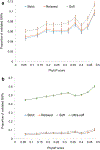
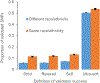
Genome wide association studies (GWASs) have identified tens of thousands of single nucleotide polymorphisms (SNPs) associated with human diseases and characteristics. A significant fraction of GWAS findings can be false positives. The gold standard for true positives is an independent validation. The goal of this study was to identify SNP features associated with validation success. Summary statistics from the Catalog of Published GWASs were used in the analysis. Since our goal was an analysis of reproducibility, we focused on the diseases/phenotypes targeted by at least 10 GWASs. GWASs were arranged in discovery-validation pairs based on the time of publication, with the discovery GWAS published before validation. We used four definitions of the validation success that differ by stringency. Associations of SNP features with validation success were consistent across the definitions. The strongest predictor of SNP validation was the level of statistical significance in the discovery GWAS. The magnitude of the effect size was associated with validation success in a non-linear manner. SNPs with risk allele frequencies in the range 30-70% showed a higher validation success rate compared to rarer or more common SNPs. Missense, 5'UTR, stop gained, and SNPs located in transcription factor binding sites had a higher validation success rate compared to intergenic, intronic and synonymous SNPs. There was a positive association between validation success and the level of evolutionary conservation of the sites. In addition, validation success was higher when discovery and validation GWASs targeted the same ethnicity. All predictors of validation success remained significant in a multivariate logistic regression model indicating their independent contribution. To conclude, we identified SNP features predicting validation success of GWAS hits. These features can be used to select SNPs for validation and downstream functional studies.
© 2021. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
Conflicts of interest/Competing interests
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

