Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis
- PMID: 34982022
- PMCID: PMC8805892
- DOI: 10.1080/21655979.2021.2019172
Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis
Abstract
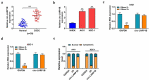
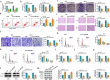
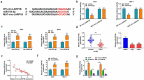
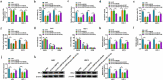
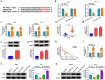
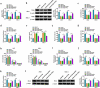
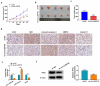
Circular RNAs (circRNAs) have shown pivotal regulatory roles in tumorigenesis and progression. Our purpose was to analyze the role of circRNA La ribonucleoprotein 1B (circ-LARP1B; hsa_circ_0070934) in cutaneous squamous cell carcinoma (CSCC) progression and its associated mechanism. Cell viability, colony formation ability, migration, and invasion were analyzed by 3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyltetrazolium bromide (MTT) assay, colony formation assay, wound healing assay, and transwell invasion assay. Flow cytometry was performed to analyze cell apoptosis and cell cycle progression. Cell glycolytic metabolism was analyzed using Glucose Uptake Colorimetric Assay kit, Lactate Assay Kit II, and ATP colorimetric Assay kit. Dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were performed to verify the interaction between microRNA-515-5p (miR-515-5p) and circ-LARP1B or TPX2 microtubule nucleation factor (TPX2). Circ-LARP1B expression was up-regulated in CSCC tissues and cell lines. Circ-LARP1B knockdown suppressed cell viability, colony formation ability, migration, invasion, cell cycle progression, and glycolysis and triggered cell apoptosis in CSCC cells. miR-515-5p was a direct target of circ-LARP1B in CSCC cells, and circ-LARP1B silencing-mediated anti-tumor effects were largely counteracted by miR-515-5p knockdown. miR-515-5p directly interacted with the 3' untranslated region (3'UTR) of TPX2. TPX2 overexpression largely overturned miR-515-5p-mediated anti-tumor effects in CSCC cells. Circ-LARP1B could up-regulate TPX2 expression by sponging miR-515-5p in CSCC cells. Circ-LARP1B knockdown suppressed tumor growth in vivo. In conclusion, circ-LARP1B contributed to CSCC progression by targeting miR-515-5p/TPX2 axis. The circ-LARP1B/miR-515-5p/TPX2 axis might provide novel therapeutic targets for CSCC patients.
Keywords: Cutaneous squamous cell carcinoma; TPX2; circ-LARP1B; miR-515-5p.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
CircRNA circ_0067772 aggravates the malignant progression of cutaneous squamous cell carcinoma by regulating miR-1238-3p/FOXG1 axis.Genes Genomics. 2021 May;43(5):491-501. doi: 10.1007/s13258-021-01074-3. Epub 2021 Mar 11. Genes Genomics. 2021. PMID: 33709381
-
Upregulated circular RNA circ_0070934 facilitates cutaneous squamous cell carcinoma cell growth and invasion by sponging miR-1238 and miR-1247-5p.Biochem Biophys Res Commun. 2019 May 28;513(2):380-385. doi: 10.1016/j.bbrc.2019.04.017. Epub 2019 Apr 6. Biochem Biophys Res Commun. 2019. PMID: 30967263
-
Circ_0068631 sponges miR-139-5p to promote the growth and metastasis of cutaneous squamous cell carcinoma by upregulating HOXB7.Skin Res Technol. 2023 Feb;29(2):e13248. doi: 10.1111/srt.13248. Skin Res Technol. 2023. PMID: 36823512 Free PMC article.
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
-
Circ_CDR1as: A circular RNA with roles in the carcinogenesis.Pathol Res Pract. 2022 Aug;236:153968. doi: 10.1016/j.prp.2022.153968. Epub 2022 Jun 1. Pathol Res Pract. 2022. PMID: 35667198 Review.
Cited by
-
Circular RNA PGPEP1 induces colorectal cancer malignancy and immune escape.Cell Cycle. 2023 Jul-Aug;22(14-16):1743-1758. doi: 10.1080/15384101.2023.2225923. Epub 2023 Jul 9. Cell Cycle. 2023. PMID: 37424115 Free PMC article.
-
LncRNA growth arrest specific transcript 5 inhibits the growth of pituitary neuroendocrine tumors via miR-27a-5p/cylindromatosis axis.Bioengineered. 2022 Apr;13(4):10274-10286. doi: 10.1080/21655979.2022.2062086. Bioengineered. 2022. PMID: 35435104 Free PMC article.
-
Circular RNA CDK6 suppresses cervical cancer proliferation and metastasis by sponging miR-449a.Bioengineered. 2022 Mar;13(3):4885-4897. doi: 10.1080/21655979.2022.2036898. Bioengineered. 2022. PMID: 35152839 Free PMC article.
-
Non-coding RNAs in skin cancers:Biological roles and molecular mechanisms.Front Pharmacol. 2022 Aug 10;13:934396. doi: 10.3389/fphar.2022.934396. eCollection 2022. Front Pharmacol. 2022. PMID: 36034860 Free PMC article. Review.
-
CircLARP1B promotes pyroptosis of high glucose-induced renal mesangial cells by regulating the miR-578/TLR4 axis.Int Urol Nephrol. 2024 Jan;56(1):283-293. doi: 10.1007/s11255-023-03672-4. Epub 2023 Jun 21. Int Urol Nephrol. 2024. PMID: 37341906
References
-
- Liu T, Lei Z, Pan Z, et al. Genetic association between p53 codon 72 polymorphism and risk of cutaneous squamous cell carcinoma. Tumour Biol. 2014;35(4):3899–3903. - PubMed
-
- Ishitsuka Y, Kawachi Y, Taguchi S, et al. Pituitary tumor-transforming gene 1 as a proliferation marker lacking prognostic value in cutaneous squamous cell carcinoma. Exp Dermatol. 2013;22(5):318–322. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
