Engineered small extracellular vesicles displaying ACE2 variants on the surface protect against SARS-CoV-2 infection
- PMID: 34982509
- PMCID: PMC8725171
- DOI: 10.1002/jev2.12179
Engineered small extracellular vesicles displaying ACE2 variants on the surface protect against SARS-CoV-2 infection
Abstract
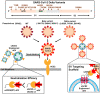
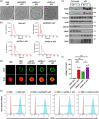
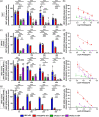
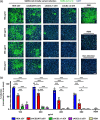
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry is mediated by the interaction of the viral spike (S) protein with angiotensin-converting enzyme 2 (ACE2) on the host cell surface. Although a clinical trial testing soluble ACE2 (sACE2) for COVID-19 is currently ongoing, our understanding of the delivery of sACE2 via small extracellular vesicles (sEVs) is still rudimentary. With excellent biocompatibility allowing for the effective delivery of molecular cargos, sEVs are broadly studied as nanoscale protein carriers. In order to exploit the potential of sEVs, we design truncated CD9 scaffolds to display sACE2 on the sEV surface as a decoy receptor for the S protein of SARS-CoV-2. Moreover, to enhance the sACE2-S binding interaction, we employ sACE2 variants. sACE2-loaded sEVs exhibit typical sEVs characteristics and bind to the S protein. Furthermore, engineered sEVs inhibit the entry of wild-type (WT), the globally dominant D614G variant, Beta (K417N-E484K-N501Y) variant, and Delta (L452R-T478K-D614G) variant SARS-CoV-2 pseudovirus, and protect against authentic SARS-CoV-2 and Delta variant infection. Of note, sACE2 variants harbouring sEVs show superior antiviral efficacy than WT sACE2 loaded sEVs. Therapeutic efficacy of the engineered sEVs against SARS-CoV-2 challenge was confirmed using K18-hACE2 mice. The current findings provide opportunities for the development of new sEVs-based antiviral therapeutics.
Keywords: COVID-19; SARS-CoV-2; beta variant; delta variant; extracellular vesicles; soluble ACE2; spike.
© 2022 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
Y.W.C. is the chief executive officer of ExoStemTech Inc. D.‐G.J. and J.H.P. are stockholders of ExoStemTech Inc. H.K.K, E.K., J.K., Y.W.C., D.‐G.J. have filed a patent application based on this work.
Figures

References
-
- Boucheix, C. , Benoit, P. , Frachet, P. , Billard, M. , Worthington, R. E. , Gagnon, J. , & Uzan, G. (1991). Molecular cloning of the CD9 antigen. A new family of cell surface proteins. The Journal of Biological Chemistry, 266, 117–122. - PubMed
-
- Cocozza, F. , Névo, N. , Piovesana, E. , Lahaye, X. , Buchrieser, J. , Schwartz, O. , Manel, N. , Tkach, M. , Théry, C. , & Martin‐Jaular, L. (2020). Extracellular vesicles containing ACE2 efficiently prevent infection by SARS‐CoV‐2 Spike protein‐containing virus. Journal of Extracellular Vesicles, 10, e12050. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NRF-2019R1A2C3011422/the National Research Foundation of Korea
- NRF-2019R1A5A2027340/the National Research Foundation of Korea
- 1525011845/the Ministry of Oceans and Fisheries' R&D project, Korea
- Korea Basic Science Institute (National Research Facilities and Equipment Center)
- 2020R1A6C101A191/the Ministry of Education
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

