The Type 2 Asthma Mediator IL-13 Inhibits Severe Acute Respiratory Syndrome Coronavirus 2 Infection of Bronchial Epithelium
- PMID: 34982656
- PMCID: PMC8990122
- DOI: 10.1165/rcmb.2021-0364OC
The Type 2 Asthma Mediator IL-13 Inhibits Severe Acute Respiratory Syndrome Coronavirus 2 Infection of Bronchial Epithelium
Abstract
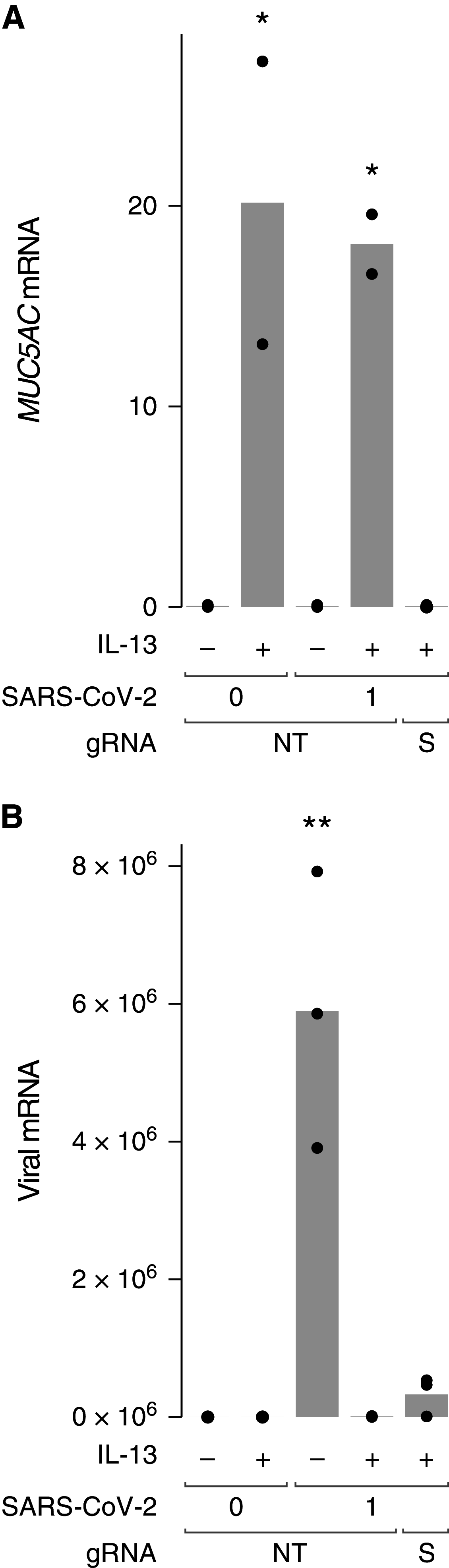
Asthma is associated with chronic changes in the airway epithelium, a key target of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Many epithelial changes, including goblet cell metaplasia, are driven by the type 2 cytokine IL-13, but the effects of IL-13 on SARS-CoV-2 infection are unknown. We found that IL-13 stimulation of differentiated human bronchial epithelial cells (HBECs) cultured at air-liquid interface reduced viral RNA recovered from SARS-CoV-2-infected cells and decreased double-stranded RNA, a marker of viral replication, to below the limit of detection in our assay. An intact mucus gel reduced SARS-CoV-2 infection of unstimulated cells, but neither a mucus gel nor SPDEF, which is required for goblet cell metaplasia, were required for the antiviral effects of IL-13. Bulk RNA sequencing revealed that IL-13 regulated 41 of 332 (12%) mRNAs encoding SARS-CoV-2-associated proteins that were detected in HBECs (>1.5-fold change; false discovery rate < 0.05). Although both IL-13 and IFN-α each inhibit SARS-CoV-2 infection, their transcriptional effects differed markedly. Single-cell RNA sequencing revealed cell type-specific differences in SARS-CoV-2-associated gene expression and IL-13 responses. Many IL-13-induced gene expression changes were seen in airway epithelium from individuals with type 2 asthma and chronic obstructive pulmonary disease. IL-13 effects on airway epithelial cells may protect individuals with type 2 asthma from COVID-19 and could lead to identification of novel strategies for reducing SARS-CoV-2 infection.
Keywords: COVID-19; IL-13; SARS-CoV-2; airway epithelium; asthma.
Figures



Update of
-
The type 2 asthma mediator IL-13 inhibits SARS-CoV-2 infection of bronchial epithelium.bioRxiv [Preprint]. 2021 Feb 25:2021.02.25.432762. doi: 10.1101/2021.02.25.432762. bioRxiv. 2021. Update in: Am J Respir Cell Mol Biol. 2022 Apr;66(4):391-401. doi: 10.1165/rcmb.2021-0364OC. PMID: 33655249 Free PMC article. Updated. Preprint.
Comment in
-
IL-13 Protects against SARS-CoV-2?Am J Respir Cell Mol Biol. 2022 Apr;66(4):351-352. doi: 10.1165/rcmb.2021-0562ED. Am J Respir Cell Mol Biol. 2022. PMID: 35085479 Free PMC article. No abstract available.
References
-
- Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, et al. HGID Lab; NIAID-USUHS Immune Response to COVID Group; COVID Clinicians; COVID-STORM Clinicians; Imagine COVID Group; French COVID Cohort Study Group; Milieu Intérieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science . 2020;370:eabd4585. - PMC - PubMed
-
- Centers for Disease Control and Prevention 2020https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/evidenc....

