The Candida albicans Cdk8-dependent phosphoproteome reveals repression of hyphal growth through a Flo8-dependent pathway
- PMID: 34982775
- PMCID: PMC8769334
- DOI: 10.1371/journal.pgen.1009622
The Candida albicans Cdk8-dependent phosphoproteome reveals repression of hyphal growth through a Flo8-dependent pathway
Abstract
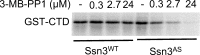
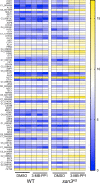
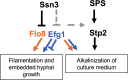
Ssn3, also known as Cdk8, is a member of the four protein Cdk8 submodule within the multi-subunit Mediator complex involved in the co-regulation of transcription. In Candida albicans, the loss of Ssn3 kinase activity affects multiple phenotypes including cellular morphology, metabolism, nutrient acquisition, immune cell interactions, and drug resistance. In these studies, we generated a strain in which Ssn3 was replaced with a functional variant of Ssn3 that can be rapidly and selectively inhibited by the ATP analog 3-MB-PP1. Consistent with ssn3 null mutant and kinase dead phenotypes, inhibition of Ssn3 kinase activity promoted hypha formation. Furthermore, the increased expression of hypha-specific genes was the strongest transcriptional signal upon inhibition of Ssn3 in transcriptomics analyses. Rapid inactivation of Ssn3 was used for phosphoproteomic studies performed to identify Ssn3 kinase substrates associated with filamentation potential. Both previously validated and novel Ssn3 targets were identified. Protein phosphorylation sites that were reduced specifically upon Ssn3 inhibition included two sites in Flo8 which is a transcription factor known to positively regulate C. albicans morphology. Mutation of the two Flo8 phosphosites (threonine 589 and serine 620) was sufficient to increase Flo8-HA levels and Flo8 dependent transcriptional and morphological changes, suggesting that Ssn3 kinase activity negatively regulates Flo8.Under embedded conditions, when ssn3Δ/Δ and efg1Δ/Δ mutants were hyperfilamentous, FLO8 was essential for hypha formation. Previous work has also shown that loss of Ssn3 activity leads to increased alkalinization of medium with amino acids. Here, we show that the ssn3Δ/Δ medium alkalinization phenotype, which is dependent on STP2, a transcription factor involved in amino acid utilization, also requires FLO8 and EFG1. Together, these data show that Ssn3 activity can modulate Flo8 and its direct and indirect interactions in different ways, and underscores the potential importance of considering Ssn3 function in the control of transcription factor activities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

