A somatic UBA2 variant preceded ETV6-RUNX1 in the concordant BCP-ALL of monozygotic twins
- PMID: 34982829
- PMCID: PMC9006272
- DOI: 10.1182/bloodadvances.2021005703
A somatic UBA2 variant preceded ETV6-RUNX1 in the concordant BCP-ALL of monozygotic twins
Abstract
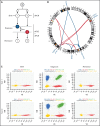
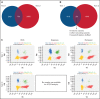
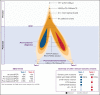
Genetic analysis of leukemic clones in monozygotic twins with concordant acute lymphoblastic leukemia (ALL) has proved a unique opportunity to gain insight into the molecular phylogenetics of leukemogenesis. Using whole-genome sequencing, we characterized constitutional and somatic single nucleotide variants/insertion-deletions (indels) and structural variants in a monozygotic twin pair with concordant ETV6-RUNX1+ B-cell precursor ALL (BCP-ALL). In addition, digital PCR (dPCR) was applied to evaluate the presence of and quantify selected somatic variants at birth, diagnosis, and remission. A shared somatic complex rearrangement involving chromosomes 11, 12, and 21 with identical fusion sequences in leukemias of both twins offered direct proof of a common clonal origin. The ETV6-RUNX1 fusion detected at diagnosis was found to originate from this complex rearrangement. A shared somatic frameshift deletion in UBA2 was also identified in diagnostic samples. In addition, each leukemia independently acquired analogous deletions of 3 genes recurrently targeted in BCP-ALLs (ETV6, ATF7IP, and RAG1/RAG2), providing evidence of a convergent clonal evolution only explained by a strong concurrent selective pressure. Quantification of the UBA2 deletion by dPCR surprisingly indicated it persisted in remission. This, for the first time to our knowledge, provided evidence of a UBA2 variant preceding the well-established initiating event ETV6-RUNX1. Further, we suggest the UBA2 deletion exerted a leukemia predisposing effect and that its essential role in Small Ubiquitin-like Modifier (SUMO) attachment (SUMOylation), regulating nearly all physiological and pathological cellular processes such as DNA-repair by nonhomologous end joining, may hold a mechanistic explanation for the predisposition.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

References
-
- Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541-1552. - PubMed

