Expression of immune response genes in human corneal epithelial cells interacting with Aspergillus flavus conidia
- PMID: 34983375
- PMCID: PMC8728928
- DOI: 10.1186/s12864-021-08218-5
Expression of immune response genes in human corneal epithelial cells interacting with Aspergillus flavus conidia
Abstract
Background: Aspergillus flavus, one of the causative agents of human fungal keratitis, can be phagocytosed by human corneal epithelial (HCE) cells and the conidia containing phagosomes mature into phagolysosomes. But the immunological responses of human corneal epithelial cells interacting with A. flavus are not clear. In this study, we report the expression of immune response related genes of HCE cells exposed to A. flavus spores using targeted transcriptomics.
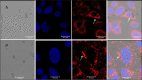
Methods: Human corneal epithelial cell line and primary cultures were grown in a six-well plate and used for coculture experiments. Internalization of the conidia was confirmed by immunofluorescence microscopy of the colocalized endosomal markers CD71 and LAMP1. Total RNA was isolated, and the quantity and quality of the isolated RNA were assessed using Qubit and Bioanalyzer. NanoString nCounter platform was used for the analysis of mRNA abundance using the Human Immunology panel. R-package and nSolver software were used for data analysis. KEGG and FunRich 3.1.3 tools were used to analyze the differentially expressed genes.
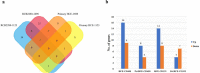
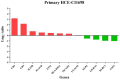
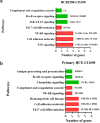
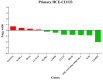
Results: Different morphotypes of conidia were observed after 6 h of coculture with human corneal epithelial cells and found to be internalized by epithelial cells. NanoString profiling showed more than 20 differentially expressed genes in immortalized human corneal epithelial cell line and more than ten differentially expressed genes in primary corneal epithelial cells. Distinct set of genes were altered in their expression in cell line and primary corneal epithelial cells. KEGG pathway analysis revealed that genes associated with TNF signaling, NF-KB signaling, and Th17 signaling were up-regulated, and genes associated with chemokine signaling and B cell receptor signaling were down regulated. FunRich pathway analysis showed that pathways such as CDC42 signaling, PI3K signaling, and Arf6 trafficking events were activated by the clinical isolates CI1123 and CI1698 in both type of cells.
Conclusions: Combining the transcript analysis data from cell lines and primary cultures, we showed the up regulation of immune defense genes in A. flavus infected cells. At the same time, chemokine signaling and B cell signaling pathways are downregulated. The variability in the expression levels in the immortalized cell line and the primary cultures is likely due to the variable epigenetic reprogramming in the immortalized cells and primary cultures in the absence of any changes in the genome. It highlights the importance of using both cell types in host-pathogen interaction studies.
Keywords: Aspergillus flavus; Corneal epithelial cells; Fungal keratitis; Immune response; NanoString analysis; Targeted transcriptomics.
© 2021. The Author(s).
Conflict of interest statement
Authors declare that they have no competing interests.
Figures










References
-
- Chang PK, Scharfenstein LL, Mack B, Ehrlich KC. Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl Environ Microbiol. 2012;78:7557–63. doi: 10.1128/AEM.01241-12. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

