Inhibition of aberrant Hif1α activation delays intervertebral disc degeneration in adult mice
- PMID: 34983922
- PMCID: PMC8727577
- DOI: 10.1038/s41413-021-00165-x
Inhibition of aberrant Hif1α activation delays intervertebral disc degeneration in adult mice
Abstract
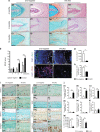
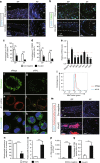
The intervertebral disc (IVD) is the largest avascular tissue. Hypoxia-inducible factors (HIFs) play essential roles in regulating cellular adaptation in the IVD under physiological conditions. Disc degeneration disease (DDD) is one of the leading causes of disability, and current therapies are ineffective. This study sought to explore the role of HIFs in DDD pathogenesis in mice. The findings of this study showed that among HIF family members, Hif1α was significantly upregulated in cartilaginous endplate (EP) and annulus fibrosus (AF) tissues from human DDD patients and two mouse models of DDD compared with controls. Conditional deletion of the E3 ubiquitin ligase Vhl in EP and AF tissues of adult mice resulted in upregulated Hif1α expression and age-dependent IVD degeneration. Aberrant Hif1α activation enhanced glycolytic metabolism and suppressed mitochondrial function. On the other hand, genetic ablation of the Hif1α gene delayed DDD pathogenesis in Vhl-deficient mice. Administration of 2-methoxyestradiol (2ME2), a selective Hif1α inhibitor, attenuated experimental IVD degeneration in mice. The findings of this study show that aberrant Hif1α activation in EP and AF tissues induces pathological changes in DDD, implying that inhibition of aberrant Hif1α activity is a potential therapeutic strategy for DDD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Balague F, Mannion AF, Pellise F, Cedraschi C. Non-specific low back pain. Lancet. 2012;379:482–491. - PubMed
-
- Andersson GB. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–585. - PubMed
-
- Kerr GJ, Veras MA, Kim MK, Seguin CA. Decoding the intervertebral disc: unravelling the complexities of cell phenotypes and pathways associated with degeneration and mechanotransduction. Semin. Cell Dev. Biol. 2017;62:94–103. - PubMed
-
- Boden SD, Davis DO, Dina TS, Patronas NJ, Wiesel SW. Abnormal magnetic-resonance scans of the lumbar spine in asymptomatic subjects. A prospective investigation. J. Bone Jt. Surg. Am. 1990;72:403–408. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

