Whole genome sequence of bacteremic Clostridium tertium in a World War I soldier, 1914
- PMID: 34984406
- PMCID: PMC8693014
- DOI: 10.1016/j.crmicr.2021.100089
Whole genome sequence of bacteremic Clostridium tertium in a World War I soldier, 1914
Abstract
Background: Dental pulp, encapsulating a blood drop, could be used to diagnose pathogen bacteraemia in archaeological materials using DNA-based techniques. We questioned the viability of such ancient pathogens preserved in ancient dental pulp.
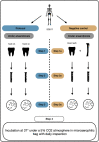
Methods: After meticulous decontamination of 32 teeth collected from 31 World War I soldiers exhumed in Spincourt, France, dental pulps were extracted and cultured under strict anaerobiosis. Colonies were identified by mass spectrometry and whole genome sequencing. Fluorescent in situ hybridisation (FISH) was used for the direct microscopic detection of pathogens of interest in the dental pulp. All the experimental procedures included negative controls, notably sediments in contact with individual SQ517 to ensure that results did not arise from contamination.
Findings: Clostridium tertium was detected by FISH in two dental pulp specimens taken from a 1914 soldier. After a two-day incubation period, both dental pulp samples grew colonies identified by mass spectrometry and genome sequencing as C. tertium; whereas negative controls remained free of C. tertium in all the observations, and no C. tertium was founded in sediments. Skeletal remains of this soldier exhibited two notches in the left tibia evocative of a cold steel wound, and a probably fatal unhealed bullet impact in the hip bone.
Interpretation: Data indicated the presence of C. tertium in the dental pulp at the time of the death of one World War I soldier, in 1914. This observation diagnosed C. tertium bacteraemia, with war wounds as the probable portal of entry for C. tertium. Our C. tertium strains ante-dated by three years, the princeps description of this deadly opportunistic pathogen.
Keywords: Clostridium tertium; Culturomics; Dental pulp; Paleomicrobiology; Soldier; WWI.
© 2021 The Authors. Published by Elsevier B.V.
Conflict of interest statement
All authors declare no competing interests.
Figures







References
-
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prijibelski A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. - PMC - PubMed
-
- Brůžek J., Santos F., Dutailly B., Murail P., Cunha E. Validation and reliability of the sex estimation of the human os coxae using freely available DSP2 software for bioarchaeology and forensic anthropology. Am. J. Phys. Anthropol. 2017;164(2):440–449. - PubMed
LinkOut - more resources
Full Text Sources

