Liquid biopsy detection of genomic alterations in pediatric brain tumors from cell-free DNA in peripheral blood, CSF, and urine
- PMID: 34984433
- PMCID: PMC9340641
- DOI: 10.1093/neuonc/noab299
Liquid biopsy detection of genomic alterations in pediatric brain tumors from cell-free DNA in peripheral blood, CSF, and urine
Abstract
Background: The ability to identify genetic alterations in cancers is essential for precision medicine; however, surgical approaches to obtain brain tumor tissue are invasive. Profiling circulating tumor DNA (ctDNA) in liquid biopsies has emerged as a promising approach to avoid invasive procedures. Here, we systematically evaluated the feasibility of profiling pediatric brain tumors using ctDNA obtained from plasma, cerebrospinal fluid (CSF), and urine.
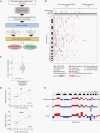
Methods: We prospectively collected 564 specimens (257 blood, 240 urine, and 67 CSF samples) from 258 patients across all histopathologies. We performed ultra-low-pass whole-genome sequencing (ULP-WGS) to assess copy number variations and estimate tumor fraction and developed a pediatric CNS tumor hybrid capture panel for deep sequencing of specific mutations and fusions.
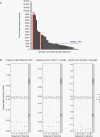
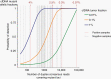
Results: ULP-WGS detected copy number alterations in 9/46 (20%) CSF, 3/230 (1.3%) plasma, and 0/153 urine samples. Sequencing detected alterations in 3/10 (30%) CSF, 2/74 (2.7%) plasma, and 0/2 urine samples. The only positive results were in high-grade tumors. However, most samples had insufficient somatic mutations (median 1, range 0-39) discoverable by the sequencing panel to provide sufficient power to detect tumor fractions of greater than 0.1%.
Conclusions: Children with brain tumors harbor very low levels of ctDNA in blood, CSF, and urine, with CSF having the most DNA detectable. Molecular profiling is feasible in a small subset of high-grade tumors. The level of clonal aberrations per genome is low in most of the tumors, posing a challenge for detection using whole-genome or even targeted sequencing methods. Substantial challenges therefore remain to genetically characterize pediatric brain tumors from liquid biopsies.
Keywords: ULP-WGS; circulating tumor DNA; hybrid capture sequencing; liquid biopsy; pediatric brain tumors.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Comment in
-
Meeting the high expectations for liquid biopsy assays for pediatric brain tumors: Progress and challenges.Neuro Oncol. 2022 Aug 1;24(8):1364-1365. doi: 10.1093/neuonc/noac083. Neuro Oncol. 2022. PMID: 35381089 Free PMC article. No abstract available.
References
-
- Zill OA, Banks KC, Fairclough SR, et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24(15):3528–3538. - PubMed
-
- Martínez-Ricarte F, Mayor R, Martíínez-Sáez E, et al. Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumor DNA from cerebrospinal fluid. Clin Cancer Res. 2018;24(12):2812–2819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

