Proteomics analysis reveals that foreign cp4-epsps gene regulates the levels of shikimate and branched pathways in genetically modified soybean line H06-698
- PMID: 34984949
- PMCID: PMC9208623
- DOI: 10.1080/21645698.2021.2000320
Proteomics analysis reveals that foreign cp4-epsps gene regulates the levels of shikimate and branched pathways in genetically modified soybean line H06-698
Abstract
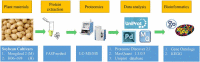
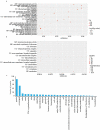
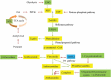
Although genetically modified (GM) glyphosate-resistant soybeans with cp4-epsps gene have been widely planted all over the world, their proteomic characteristics are not very clear. In this study, the soybean seeds of a GM soybean line H06-698 (H) with cp4-epsps gene and its non-transgenic counterpart Mengdou12 (M), which were collected from two experiment fields in two years and used as 4 sample groups, were analyzed with label-free proteomics technique. A total of 1706 proteins were identified quantitatively by label-free quantification, and a total of 293 proteins were detected as common differential abundance proteins (DAPs, FC is not less than 1.5) both in two groups or more. Functional enrichment analysis of common DAPs identified from four groups, shows that most up-regulated proteins were clustered into stress response, carbon and energy metabolism, and genetic information processing. Further documentary analysis shows that 15 proteins play important roles in shikimate pathways, reactive oxygen species (ROS) and stress response. These results indicated that the change of protein abundance in different samples were affected by various factors, but except shikimate and branched pathways related proteins, only ROS and stress-related proteins were found to be stably regulated by cp4-epsps gene, and no unexpected and safety-related proteins such as antinutritional factors, allergenic proteins, and toxic proteins were found as DAPs. The influence of foreign genes in genetically modified plants is worthy of attention and this work provides new clues for exploring the regulated proteins and pathways in GM plants.
Keywords: Cp4-epsps gene; genetically modified; label-free proteomics; shikimate pathway; soybean.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
A simple electrochemical immunosensor for sensitive detection of transgenic soybean protein CP4-EPSPS in seeds.Talanta. 2022 Jan 15;237:122910. doi: 10.1016/j.talanta.2021.122910. Epub 2021 Oct 1. Talanta. 2022. PMID: 34736647
-
The combination of quantitative PCR and western blot detecting CP4-EPSPS component in Roundup Ready soy plant tissues and commercial soy-related foodstuffs.J Food Sci. 2012 Jun;77(6):C603-8. doi: 10.1111/j.1750-3841.2012.02718.x. Epub 2012 May 16. J Food Sci. 2012. PMID: 22591269
-
Variability of CP4 EPSPS expression in genetically engineered soybean (Glycine max L. Merrill).Transgenic Res. 2018 Dec;27(6):511-524. doi: 10.1007/s11248-018-0092-z. Epub 2018 Sep 1. Transgenic Res. 2018. PMID: 30173346 Free PMC article.
-
Expression of CP4 EPSPS in microspores and tapetum cells of cotton (Gossypium hirsutum) is critical for male reproductive development in response to late-stage glyphosate applications.Plant Biotechnol J. 2006 Sep;4(5):477-87. doi: 10.1111/j.1467-7652.2006.00203.x. Plant Biotechnol J. 2006. PMID: 17309724 Review.
-
A review of the environmental safety of the CP4 EPSPS protein.Environ Biosafety Res. 2011 Jan-Mar;10(1):5-25. doi: 10.1051/ebr:2012001. Epub 2012 Apr 30. Environ Biosafety Res. 2011. PMID: 22541883 Review. No abstract available.
Cited by
-
Nocardioides imazamoxiresistens sp. nov. Isolated from the Activated Sludge.Curr Microbiol. 2024 Jun 7;81(7):214. doi: 10.1007/s00284-024-03731-8. Curr Microbiol. 2024. PMID: 38849626
-
Insights into the effects of transgenic glyphosate-resistant semiwild soybean on soil microbial diversity.Sci Rep. 2024 Dec 30;14(1):32017. doi: 10.1038/s41598-024-83676-x. Sci Rep. 2024. PMID: 39738641 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
