Activation by cleavage of the epithelial Na+ channel α and γ subunits independently coevolved with the vertebrate terrestrial migration
- PMID: 34984981
- PMCID: PMC8791634
- DOI: 10.7554/eLife.75796
Activation by cleavage of the epithelial Na+ channel α and γ subunits independently coevolved with the vertebrate terrestrial migration
Abstract
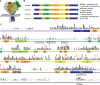
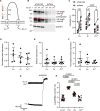
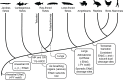
Vertebrates evolved mechanisms for sodium conservation and gas exchange in conjunction with migration from aquatic to terrestrial habitats. Epithelial Na+ channel (ENaC) function is critical to systems responsible for extracellular fluid homeostasis and gas exchange. ENaC is activated by cleavage at multiple specific extracellular polybasic sites, releasing inhibitory tracts from the channel's α and γ subunits. We found that proximal and distal polybasic tracts in ENaC subunits coevolved, consistent with the dual cleavage requirement for activation observed in mammals. Polybasic tract pairs evolved with the terrestrial migration and the appearance of lungs, coincident with the ENaC activator aldosterone, and appeared independently in the α and γ subunits. In summary, sites within ENaC for protease activation developed in vertebrates when renal Na+ conservation and alveolar gas exchange were required for terrestrial survival.
Keywords: E. calabaricus; N. forsteri; epithelial Na+ channel; evolutionary biology; furin; proteolytic activation; vertebrate land invasion; xenopus.
© 2022, Wang et al.
Conflict of interest statement
XW, DB, CG, NC, OK No competing interests declared
Figures


Similar articles
-
Influence of proteolytic cleavage of ENaC's γ subunit upon Na+ and K+ handling.Am J Physiol Renal Physiol. 2024 Jun 1;326(6):F1066-F1077. doi: 10.1152/ajprenal.00027.2024. Epub 2024 Apr 18. Am J Physiol Renal Physiol. 2024. PMID: 38634134 Free PMC article.
-
Transmembrane serine protease 2 (TMPRSS2) proteolytically activates the epithelial sodium channel (ENaC) by cleaving the channel's γ-subunit.J Biol Chem. 2022 Jun;298(6):102004. doi: 10.1016/j.jbc.2022.102004. Epub 2022 Apr 30. J Biol Chem. 2022. PMID: 35504352 Free PMC article.
-
TMPRSS4-dependent activation of the epithelial sodium channel requires cleavage of the γ-subunit distal to the furin cleavage site.Am J Physiol Renal Physiol. 2012 Jan 1;302(1):F1-8. doi: 10.1152/ajprenal.00330.2011. Epub 2011 Oct 12. Am J Physiol Renal Physiol. 2012. PMID: 21993886 Free PMC article.
-
Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases.Gene. 2016 Apr 1;579(2):95-132. doi: 10.1016/j.gene.2015.12.061. Epub 2016 Jan 7. Gene. 2016. PMID: 26772908 Free PMC article. Review.
-
Evolution of epithelial sodium channels: current concepts and hypotheses.Am J Physiol Regul Integr Comp Physiol. 2020 Oct 1;319(4):R387-R400. doi: 10.1152/ajpregu.00144.2020. Epub 2020 Aug 12. Am J Physiol Regul Integr Comp Physiol. 2020. PMID: 32783689 Review.
Cited by
-
Loss of the alpha subunit distal furin cleavage site blunts ENaC activation following Na+ restriction.J Physiol. 2024 Sep;602(17):4309-4326. doi: 10.1113/JP286559. Epub 2024 Aug 28. J Physiol. 2024. PMID: 39196791 Free PMC article.
-
MicroRNA-19 is regulated by aldosterone in a sex-specific manner to alter kidney sodium transport.Am J Physiol Cell Physiol. 2024 Jan 1;326(1):C282-C293. doi: 10.1152/ajpcell.00385.2023. Epub 2023 Dec 4. Am J Physiol Cell Physiol. 2024. PMID: 38047299 Free PMC article.
-
Varying Selection Pressure for a Na+ Sensing Site in Epithelial Na+ Channel Subunits Reflect Divergent Roles in Na+ Homeostasis.Mol Biol Evol. 2024 Aug 2;41(8):msae162. doi: 10.1093/molbev/msae162. Mol Biol Evol. 2024. PMID: 39101592 Free PMC article.
-
Epithelial Na + Channels Function as Extracellular Sensors.Compr Physiol. 2024 Mar 29;14(2):1-41. doi: 10.1002/cphy.c230015. Compr Physiol. 2024. PMID: 39109974 Free PMC article. Review.
-
The evolutionary path of the epithelial sodium channel δ-subunit in Cetartiodactyla points to a role in sodium sensing.bioRxiv [Preprint]. 2024 Nov 19:2024.11.18.623996. doi: 10.1101/2024.11.18.623996. bioRxiv. 2024. Update in: Commun Biol. 2025 Jul 4;8(1):1004. doi: 10.1038/s42003-025-08436-7. PMID: 39605611 Free PMC article. Updated. Preprint.
References
-
- Bertrand S, Camasses A, Paris M, Holland ND, Escriva H. Phylogenetic analysis of Amphioxus genes of the proprotein convertase family, including aPC6C, a marker of epithelial fusions during embryology. International Journal of Biological Sciences. 2006;2:125–132. doi: 10.7150/ijbs.2.125. - DOI - PMC - PubMed
-
- Brito PM, Meunier FJ, Clément G, Geffard-kuriyama D. The histological structure of the calcified lung of the fossil coelacanth Axelrodichthys araripensis (Actinistia: Mawsoniidae) Palaeontology. 2010;53:1281–1290. doi: 10.1111/j.1475-4983.2010.01015.x. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

