Developing an Amplification Refractory Mutation System-Quantitative Reverse Transcription-PCR Assay for Rapid and Sensitive Screening of SARS-CoV-2 Variants of Concern
- PMID: 34985323
- PMCID: PMC8729772
- DOI: 10.1128/spectrum.01438-21
Developing an Amplification Refractory Mutation System-Quantitative Reverse Transcription-PCR Assay for Rapid and Sensitive Screening of SARS-CoV-2 Variants of Concern
Abstract
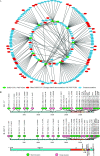
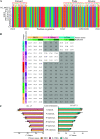
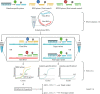
With the emergence and wide spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs), such as the Delta variant (B.1.617.2 lineage and AY sublineage), it is important to track VOCs for sourcing of transmission. Currently, whole-genome sequencing is commonly used for detecting VOCs, but this is limited by the high costs of reagents and sophisticated sequencers. In this study, common mutations in the genomes of SARS-CoV-2 VOCs were identified by analyzing more than 1 million SARS-CoV-2 genomes from public data. Among them, mutations C1709A (a change of C to A at position 1709) and C56G, respectively, were found in more than 99% of the genomes of Alpha and Delta variants and were specific to them. Then, a method using the amplification refractory mutation system combined with quantitative reverse transcription-PCR (ARMS-RT-qPCR) based on the two mutations was developed for identifying both VOCs. The assay can detect as little as 1 copy/μL of the VOCs, and the results for identifying Alpha and Delta variants in clinical samples by the ARMS-RT-qPCR assay showed 100% agreement with the results using sequencing-based methods. The whole assay can be completed in 2.5 h using commercial fluorescent PCR instruments. Therefore, the ARMS-RT-qPCR assay could be used for screening the two highly concerning variants Alpha and Delta by normal PCR laboratories in airports and in hospitals and other health-related organizations. Additionally, based on the unique mutations identified by the genomic analysis, similar molecular assays can be developed for rapid identification of other VOCs. IMPORTANCE The current stage of the pandemic, led by SARS-CoV-2 variants of concern (VOCs), underscores the necessity to develop a cost-effective and rapid molecular diagnosis assay to differentiate the VOCs. In this study, over 1 million SARS-CoV-2 genomic sequences of high quality from GISAID were analyzed and a network of the common mutations of the lineages was constructed. The conserved unique mutations specific for SARS-CoV-2 VOCs were found. Then, ARMS-RT-qPCR assays based on the two unique mutations of the Alpha and Delta variants were developed for the detection of the two VOCs. Application of the assay in clinical samples demonstrated that the current method is a convenient, cost-effective, and rapid way to screen the target SARS-CoV-2 VOCs.
Keywords: ARMS-RT-qPCR; SARS-CoV-2 variants of concern; conserved unique mutation; rapid; sensitive.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Emergency SARS-CoV-2 Variants of Concern: Novel Multiplex Real-Time RT-PCR Assay for Rapid Detection and Surveillance.Microbiol Spectr. 2022 Feb 23;10(1):e0251321. doi: 10.1128/spectrum.02513-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196812 Free PMC article.
-
Real-Time RT-PCR Allelic Discrimination Assay for Detection of N501Y Mutation in the Spike Protein of SARS-CoV-2 Associated with B.1.1.7 Variant of Concern.Microbiol Spectr. 2022 Feb 23;10(1):e0068121. doi: 10.1128/spectrum.00681-21. Epub 2022 Feb 16. Microbiol Spectr. 2022. PMID: 35170989 Free PMC article.
-
CRISPR-Cas12a-Based Detection for the Major SARS-CoV-2 Variants of Concern.Microbiol Spectr. 2021 Dec 22;9(3):e0101721. doi: 10.1128/Spectrum.01017-21. Epub 2021 Nov 17. Microbiol Spectr. 2021. PMID: 34787487 Free PMC article.
-
Molecular biology of SARS-CoV-2 and techniques of diagnosis and surveillance.Adv Clin Chem. 2024;118:35-85. doi: 10.1016/bs.acc.2023.11.003. Epub 2023 Dec 14. Adv Clin Chem. 2024. PMID: 38280807 Review.
-
An Update on Detection Technologies for SARS-CoV-2 Variants of Concern.Viruses. 2022 Oct 22;14(11):2324. doi: 10.3390/v14112324. Viruses. 2022. PMID: 36366421 Free PMC article. Review.
Cited by
-
Rapid assays of SARS-CoV-2 virus and noble biosensors by nanomaterials.Nano Converg. 2024 Jan 8;11(1):2. doi: 10.1186/s40580-023-00408-z. Nano Converg. 2024. PMID: 38190075 Free PMC article. Review.
-
Accessible and Adaptable Multiplexed Real-Time PCR Approaches to Identify SARS-CoV-2 Variants of Concern.Microbiol Spectr. 2022 Oct 26;10(5):e0322222. doi: 10.1128/spectrum.03222-22. Epub 2022 Sep 15. Microbiol Spectr. 2022. PMID: 36106882 Free PMC article.
-
Identification of potential inhibitors of omicron variant of SARS-Cov-2 RBD based virtual screening, MD simulation, and DFT.Front Chem. 2022 Dec 8;10:1063374. doi: 10.3389/fchem.2022.1063374. eCollection 2022. Front Chem. 2022. PMID: 36569957 Free PMC article.
-
Discrimination of SARS-CoV-2 omicron variant and its lineages by rapid detection of immune-escape mutations in spike protein RBD using asymmetric PCR-based melting curve analysis.Virol J. 2023 Aug 25;20(1):192. doi: 10.1186/s12985-023-02137-5. Virol J. 2023. PMID: 37626353 Free PMC article.
-
[Rapid detection and genotyping of SARS-CoV-2 Omicron BA.4/5 variants using a RT-PCR and CRISPR-Cas12a-based assay].Nan Fang Yi Ke Da Xue Xue Bao. 2023 Apr 20;43(4):516-526. doi: 10.12122/j.issn.1673-4254.2023.04.03. Nan Fang Yi Ke Da Xue Xue Bao. 2023. PMID: 37202186 Free PMC article. Chinese.
References
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Foley B, Giorgi EE, Bhattacharya T, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, LaBranche CC, Montefiori DC, Sheffield COVID-19 Genomics Group. 2020. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. doi:10.1101/2020.04.29.069054. - DOI
-
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi PY. 2021. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592:116–121. doi:10.1038/s41586-020-2895-3. - DOI - PMC - PubMed
-
- Bal A, Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, Regue H, Semanas Q, d’Aubarede C, Billaud G, Laurent F, Gonzalez C, Mekki Y, Valette M, Bouscambert M, Gaudy-Graffin C, Lina B, Morfin F, Josset L, COVID-Diagnosis HCL Study Group. 2021. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill 26:2100008. doi:10.2807/1560-7917.ES.2021.26.3.2100008. - DOI - PMC - PubMed
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. doi:10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

