Virus-Like Particles Containing the E2 Core Domain of Hepatitis C Virus Generate Broadly Neutralizing Antibodies in Guinea Pigs
- PMID: 34986001
- PMCID: PMC8906423
- DOI: 10.1128/JVI.01675-21
Virus-Like Particles Containing the E2 Core Domain of Hepatitis C Virus Generate Broadly Neutralizing Antibodies in Guinea Pigs
Abstract
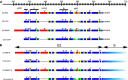
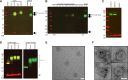
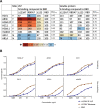
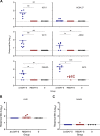
A vaccine to prevent hepatitis C virus (HCV) infection is urgently needed for use alongside direct-acting antiviral drugs to achieve elimination targets. We have previously shown that a soluble recombinant form of the glycoprotein E2 ectodomain (residues 384 to 661) that lacks three variable regions (Δ123) is able to elicit a higher titer of broadly neutralizing antibodies (bNAbs) than the parental form (receptor-binding domain [RBD]). In this study, we engineered a viral nanoparticle that displays HCV glycoprotein E2 on a duck hepatitis B virus (DHBV) small surface antigen (S) scaffold. Four variants of E2-S virus-like particles (VLPs) were constructed: Δ123-S, RBD-S, Δ123A7-S, and RBDA7-S; in the last two, 7 cysteines were replaced with alanines. While all four E2-S variant VLPs display E2 as a surface antigen, the Δ123A7-S and RBDA7-S VLPs were the most efficiently secreted from transfected mammalian cells and displayed epitopes recognized by cross-genotype broadly neutralizing monoclonal antibodies (bNMAbs). Both Δ123A7-S and RBDA7-S VLPs were immunogenic in guinea pigs, generating high titers of antibodies reactive to native E2 and able to prevent the interaction between E2 and the cellular receptor CD81. Four out of eight animals immunized with Δ123A7-S elicited neutralizing antibodies (NAbs), with three of those animals generating bNAbs against 7 genotypes. Immune serum generated by animals with NAbs mapped to major neutralization epitopes located at residues 412 to 420 (epitope I) and antigenic region 3. VLPs that display E2 glycoproteins represent a promising vaccine platform for HCV and could be adapted to large-scale manufacturing in yeast systems. IMPORTANCE There is currently no vaccine to prevent hepatitis C virus infection, which affects more than 71 million people globally and is a leading cause of progressive liver disease, including cirrhosis and cancer. Broadly neutralizing antibodies that recognize the E2 envelope glycoprotein can protect against heterologous viral infection and correlate with viral clearance in humans. However, broadly neutralizing antibodies are difficult to generate due to conformational flexibility of the E2 protein and epitope occlusion. Here, we show that a VLP vaccine using the duck hepatitis B virus S antigen fused to HCV glycoprotein E2 assembles into virus-like particles that display epitopes recognized by broadly neutralizing antibodies and elicit such antibodies in guinea pigs. This platform represents a novel HCV vaccine candidate amenable to large-scale manufacture at low cost.
Keywords: glycoprotein E2; hepatitis C virus; vaccine; virus-like particle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

