Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea
- PMID: 34986141
- PMCID: PMC9012536
- DOI: 10.1371/journal.pbio.3001508
Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea
Abstract
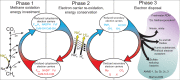
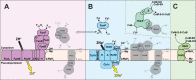
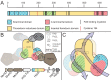
The anaerobic oxidation of methane coupled to sulfate reduction is a microbially mediated process requiring a syntrophic partnership between anaerobic methanotrophic (ANME) archaea and sulfate-reducing bacteria (SRB). Based on genome taxonomy, ANME lineages are polyphyletic within the phylum Halobacterota, none of which have been isolated in pure culture. Here, we reconstruct 28 ANME genomes from environmental metagenomes and flow sorted syntrophic consortia. Together with a reanalysis of previously published datasets, these genomes enable a comparative analysis of all marine ANME clades. We review the genomic features that separate ANME from their methanogenic relatives and identify what differentiates ANME clades. Large multiheme cytochromes and bioenergetic complexes predicted to be involved in novel electron bifurcation reactions are well distributed and conserved in the ANME archaea, while significant variations in the anabolic C1 pathways exists between clades. Our analysis raises the possibility that methylotrophic methanogenesis may have evolved from a methanotrophic ancestor.
Conflict of interest statement
S.J.H. is a co-founder of Koonkie Inc., a bioinformatics consulting company that designs and provides scalable algorithmic and data analytics solutions in the cloud.
Figures