A purine metabolic checkpoint that prevents autoimmunity and autoinflammation
- PMID: 34986329
- PMCID: PMC8730334
- DOI: 10.1016/j.cmet.2021.12.009
A purine metabolic checkpoint that prevents autoimmunity and autoinflammation
Abstract
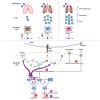
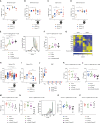
Still's disease, the paradigm of autoinflammation-cum-autoimmunity, predisposes for a cytokine storm with excessive T lymphocyte activation upon viral infection. Loss of function of the purine nucleoside enzyme FAMIN is the sole known cause for monogenic Still's disease. Here we discovered that a FAMIN-enabled purine metabolon in dendritic cells (DCs) restrains CD4+ and CD8+ T cell priming. DCs with absent FAMIN activity prime for enhanced antigen-specific cytotoxicity, IFNγ secretion, and T cell expansion, resulting in excessive influenza A virus-specific responses. Enhanced priming is already manifest with hypomorphic FAMIN-I254V, for which ∼6% of mankind is homozygous. FAMIN controls membrane trafficking and restrains antigen presentation in an NADH/NAD+-dependent manner by balancing flux through adenine-guanine nucleotide interconversion cycles. FAMIN additionally converts hypoxanthine into inosine, which DCs release to dampen T cell activation. Compromised FAMIN consequently enhances immunosurveillance of syngeneic tumors. FAMIN is a biochemical checkpoint that protects against excessive antiviral T cell responses, autoimmunity, and autoinflammation.
Keywords: NADH/NAD(+) reductive stress; T cell priming; autoimmunity; dendritic cells; membrane trafficking; purine nucleotide cycle.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The University of Cambridge has filed patent applications relating to this work. The authors declare no other competing financial interests.
Figures






References
-
- Admyre T., Amrot-Fors L., Andersson M., Bauer M., Bjursell M., Drmota T., Hallen S., Hartleib-Geschwindner J., Lindmark B., Liu J., et al. Inhibition of AMP deaminase activity does not improve glucose control in rodent models of insulin resistance or diabetes. Chem. Biol. 2014;21:1486–1496. - PubMed
-
- Al-Mayouf S.M., Almutairi A., Albrawi S., Fathalla B.M., Alzyoud R., AlEnazi A., Abu-Shukair M., Alwahadneh A., Alsonbul A., Zlenti M., et al. for Pediatric Arab Rheumatology Group (PRAG) Pattern and diagnostic evaluation of systemic autoinflammatory diseases other than familial Mediterranean fever among Arab children: a multicenter study from the Pediatric Rheumatology Arab Group (PRAG) Rheumatol. Int. 2020;40:49–56. - PubMed
-
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 103077/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- J 4396/FWF_/Austrian Science Fund FWF/Austria
- 102163/B/13/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_00014/5/MRC_/Medical Research Council/United Kingdom
- 222056/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 105920/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- 106260/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- 216630/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- R01 DK088199/DK/NIDDK NIH HHS/United States
- 222497/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- R01 DK051362/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

