Emergence of methicillin resistance predates the clinical use of antibiotics
- PMID: 34987223
- PMCID: PMC8810379
- DOI: 10.1038/s41586-021-04265-w
Emergence of methicillin resistance predates the clinical use of antibiotics
Abstract
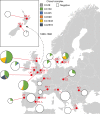
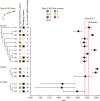
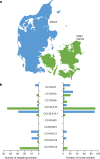
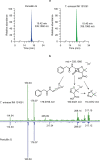
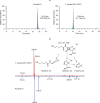
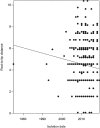
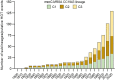
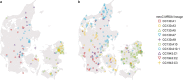
The discovery of antibiotics more than 80 years ago has led to considerable improvements in human and animal health. Although antibiotic resistance in environmental bacteria is ancient, resistance in human pathogens is thought to be a modern phenomenon that is driven by the clinical use of antibiotics1. Here we show that particular lineages of methicillin-resistant Staphylococcus aureus-a notorious human pathogen-appeared in European hedgehogs in the pre-antibiotic era. Subsequently, these lineages spread within the local hedgehog populations and between hedgehogs and secondary hosts, including livestock and humans. We also demonstrate that the hedgehog dermatophyte Trichophyton erinacei produces two β-lactam antibiotics that provide a natural selective environment in which methicillin-resistant S. aureus isolates have an advantage over susceptible isolates. Together, these results suggest that methicillin resistance emerged in the pre-antibiotic era as a co-evolutionary adaptation of S. aureus to the colonization of dermatophyte-infected hedgehogs. The evolution of clinically relevant antibiotic-resistance genes in wild animals and the connectivity of natural, agricultural and human ecosystems demonstrate that the use of a One Health approach is critical for our understanding and management of antibiotic resistance, which is one of the biggest threats to global health, food security and development.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Tracing the origins of antibiotic resistance.Nat Med. 2022 Apr;28(4):638-640. doi: 10.1038/s41591-022-01752-z. Nat Med. 2022. PMID: 35318464 No abstract available.
References
-
- European Centre for Disease Prevention and Control, European Medicines Agencies. The Bacterial Challenge: Time to React. A Call to Narrow the Gap Between Multidrug-Resistant Bacteria in the EU and the Development of New Antibacterial Agentshttps://ecdc.europa.eu/sites/portal/files/media/en/publications/Publicat... (2009).
-
- Jevons MP. “Celbenin”—resistant Staphylococci. Br. Med. J. 1961;1:124–125.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

