Whole Genome Sequencing of Staphylococci Isolated From Bovine Milk Samples
- PMID: 34987483
- PMCID: PMC8721127
- DOI: 10.3389/fmicb.2021.715851
Whole Genome Sequencing of Staphylococci Isolated From Bovine Milk Samples
Abstract
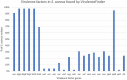
Staphylococci are among the commonly isolated bacteria from intramammary infections in bovines, where Staphylococcus aureus is the most studied species. This species carries a variety of virulence genes, contributing to bacterial survival and spread. Less is known about non-aureus staphylococci (NAS) and their range of virulence genes and mechanisms, but they are the most frequently isolated bacteria from bovine milk. Staphylococci can also carry a range of antimicrobial resistance genes, complicating treatment of the infections they cause. We used Illumina sequencing to whole genome sequence 93 staphylococcal isolates selected from a collection of staphylococcal isolates; 45 S. aureus isolates and 48 NAS isolates from 16 different species, determining their content of antimicrobial resistance genes and virulence genes. Antimicrobial resistance genes were frequently observed in the NAS species as a group compared to S. aureus. However, the lincosamide resistance gene lnuA and penicillin resistance gene blaZ were frequently identified in NAS, as well as a small number of S. aureus. The erm genes conferring macrolide resistance were also identified in several NAS isolates and in a small number of S. aureus isolates. In most S. aureus isolates, no antimicrobial resistance genes were detected, but in five S. aureus isolates three to six resistance genes were identified and all five of these carried the mecA gene. Virulence genes were more frequently identified in S. aureus, which contained on average five times more virulence genes compared to NAS. Among the NAS species there were also differences in content of virulence genes, such as S. chromogenes with a higher average number of virulence genes. By determining the content of a large selection of virulence genes and antimicrobial resistance genes in S. aureus and 16 different NAS species our results contribute with knowledge regarding the genetic basis for virulence and antimicrobial resistance in bovine staphylococci, especially the less studied NAS. The results can create a broader basis for further research into the virulence mechanisms of this important group of bacteria in bovine intramammary infections.
Keywords: Staphylococcus aureus; antimicrobial resistance (AMR) genes; bovine; non-aureus staphylococci; virulence genes; whole genome sequencing.
Copyright © 2021 Fergestad, Touzain, De Vliegher, De Visscher, Thiry, Ngassam Tchamba, Mainil, L’Abee-Lund, Blanchard and Wasteson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Aarestrup F. M., Larsen H. D., Eriksen N. H., Elsberg C. S., Jensen N. E. (1999). Frequency of alpha- and beta-haemolysin in Staphylococcus aureus of bovine and human origin. A comparison between pheno- and genotype and variation in phenotypic expression. Apmis 107 425–430. - PubMed
-
- Anthonisen I. L., Sunde M., Steinum T. M., Sidhu M. S., Sørum H. (2002). Organization of the antiseptic resistance gene qacA and Tn552-related beta-lactamase genes in multidrug- resistant Staphylococcus haemolyticus strains of animal and human origins. Antimicrob. Agents Chemother. 46 3606–3612. 10.1128/aac.46.11.3606-3612.2002 - DOI - PMC - PubMed
-
- Antignac A., Tomasz A. (2009). Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob. Agents Chemother. 53 435–441. 10.1128/aac.01099-08 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

