Adaptive Immune Response Signaling Is Suppressed in Ly6Chigh Monocyte but Upregulated in Monocyte Subsets of ApoE-/- Mice - Functional Implication in Atherosclerosis
- PMID: 34987524
- PMCID: PMC8721109
- DOI: 10.3389/fimmu.2021.809208
Adaptive Immune Response Signaling Is Suppressed in Ly6Chigh Monocyte but Upregulated in Monocyte Subsets of ApoE-/- Mice - Functional Implication in Atherosclerosis
Abstract
Rationale: Inflammatory monocyte (MC) subset differentiation is a major feature in tissue inflammatory and atherosclerosis. The underlying molecular mechanism remains unclear.
Objective: This study aims to explore molecule targets and signaling which determinate immunological features in MC subsets.
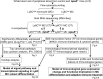
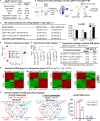
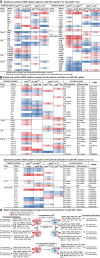
Methods and results: Blood Ly6Chigh and Ly6Clow MC subsets from control and ApoE-/- mice were isolated by flow cytometry sorting and subjected for bulk high-throughput RNA-sequencing. Intensive bioinformatic studies were performed by analyzing transcriptome through four pairs of comparisons: A) Ly6Chigh vs Ly6Clow in control mice; B) Ly6Chigh vs Ly6Clow in ApoE-/- mice; C) ApoE-/- Ly6Chigh vs control Ly6Chigh MC; D) ApoE-/- Ly6Clow vs control Ly6Clow MC. A total of 80 canonical pathways and 16 enriched pathways were recognized by top-down analysis using IPA and GSEA software, and further used for overlapping analysis. Immunological features and signaling were assessed on four selected functional groups, including MHCII, immune checkpoint, cytokine, and transcription factor (TF). Among the total 14578 significantly differentially expressed (SDE) genes identified though above four comparison, 1051 TF and 348 immunological genes were discovered. SDE immunological genes were matched with corresponding upstream SDE TF by IPA upstream analysis. Fourteen potential transcriptional axes were recognized to modulate immunological features in the Ly6C MC subset. Based on an intensive literature search, we found that the identified SDE immune checkpoint genes in Ly6Chigh MC are associated with pro-inflammatory/atherogenic balance function. Immune checkpoint genes GITR, CTLA4, and CD96 were upregulated in Ly6Clow MC from all mice and presented anti-inflammatory/atherogenic features. Six cytokine genes, including Ccl2, Tnfsf14, Il1rn, Cxcl10, Ccl9, and Cxcl2, were upregulated in Ly6Chigh MC from all mice and associated with pro-inflammatory/atherogenic feature. Cytokine receptor gene Il12rb2, Il1r1, Il27ra, Il5ra, Ngfr, Ccr7, and Cxcr5 were upregulated in Ly6Clow MC from all mice and presented anti-inflammatory/atherogenic features. MHCII genes (H2-Oa, H2-DMb2, H2-Ob, H2-Eb2, H2-Eb1, H2-Aa, and Cd74) were elevated in Ly6Clow MC from all mice. ApoE-/- augmented pro-atherogenic/inflammatory and antigen-presenting cells (APC) feature in both subsets due to elevated expression of cytokine genes (Cxcl11, Cntf, Il24, Xcl, Ccr5, Mpl, and Acvr2a) and MHCII gene (H2-Aa and H2-Ea-ps). Finally, we modeled immunological gene expression changes and functional implications in MC differentiation and adaptive immune response for MC subsets from control and ApoE-/- mice.
Conclusions: Ly6Chigh MC presented pro-inflammatory/atherogenic features and lower APC potential. Ly6Clow MC displayed anti-inflammatory/atherogenic features and higher APC potential. ApoE-/- confers upon both subsets with augmented pro-atherogenic/inflammatory function and APC potential.
Keywords: ApoE; Ly6C MC; adaptive immune response; atherosclerosis; inflammatory.
Copyright © 2021 Yang, Wu, Sun, Fang, Liu, Ji, Park, Qin, Yang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Immunological Feature and Transcriptional Signaling of Ly6C Monocyte Subsets From Transcriptome Analysis in Control and Hyperhomocysteinemic Mice.Front Immunol. 2021 Feb 25;12:632333. doi: 10.3389/fimmu.2021.632333. eCollection 2021. Front Immunol. 2021. PMID: 33717169 Free PMC article.
-
Map3k8 Modulates Monocyte State and Atherogenesis in ApoE-/- Mice.Arterioscler Thromb Vasc Biol. 2017 Feb;37(2):237-246. doi: 10.1161/ATVBAHA.116.308528. Epub 2016 Nov 17. Arterioscler Thromb Vasc Biol. 2017. PMID: 27856455
-
Suppressor of Cytokine Signaling 1 is Involved in Gene Regulation Which Controls the Survival of Ly6Clow Monocytes in Mice.Cell Physiol Biochem. 2019;52(2):336-353. doi: 10.33594/000000024. Epub 2019 Feb 28. Cell Physiol Biochem. 2019. PMID: 30816678
-
Metabolism-associated danger signal-induced immune response and reverse immune checkpoint-activated CD40+ monocyte differentiation.J Hematol Oncol. 2017 Jul 24;10(1):141. doi: 10.1186/s13045-017-0504-1. J Hematol Oncol. 2017. PMID: 28738836 Free PMC article. Review.
-
Macrophages heterogeneity in atherosclerosis - implications for therapy.J Cell Mol Med. 2010 Aug;14(8):2055-65. doi: 10.1111/j.1582-4934.2010.01121.x. Epub 2010 Jul 12. J Cell Mol Med. 2010. PMID: 20629993 Free PMC article. Review.
Cited by
-
Nobiletin restores the intestinal barrier of HFD-induced obese mice by promoting MHC-II expression and lipid metabolism.Mol Med. 2025 Jan 26;31(1):26. doi: 10.1186/s10020-025-01072-1. Mol Med. 2025. PMID: 39865231 Free PMC article.
-
The Dual Role of Innate Immune Response in Acetaminophen-Induced Liver Injury.Biology (Basel). 2022 Jul 14;11(7):1057. doi: 10.3390/biology11071057. Biology (Basel). 2022. PMID: 36101435 Free PMC article. Review.
-
Comprehensive analysis of human monocyte subsets using full-spectrum flow cytometry and hierarchical marker clustering.Front Immunol. 2024 Apr 29;15:1405249. doi: 10.3389/fimmu.2024.1405249. eCollection 2024. Front Immunol. 2024. PMID: 38742110 Free PMC article.
-
Chemokine Ligands and Receptors Regulate Macrophage Polarization in Atherosclerosis: A Comprehensive Database Mining Study.CJC Open. 2024 Nov 26;7(3):310-324. doi: 10.1016/j.cjco.2024.11.018. eCollection 2025 Mar. CJC Open. 2024. PMID: 40182401 Free PMC article.
-
CD4-Derived Double-Negative T Cells Ameliorate Alzheimer's Disease-Like Phenotypes in the 5×FAD Mouse Model.CNS Neurosci Ther. 2025 Jan;31(1):e70187. doi: 10.1111/cns.70187. CNS Neurosci Ther. 2025. PMID: 39844773 Free PMC article.
References
-
- Fang P, Li X, Shan H, Saredy JJ, Cueto R, Xia J, et al. . Ly6c(+) Inflammatory Monocyte Differentiation Partially Mediates Hyperhomocysteinemia-Induced Vascular Dysfunction in Type 2 Diabetic Db/Db Mice. Arterioscler Thromb Vasc Biol (2019) 39:2097–119. doi: 10.1161/ATVBAHA.119.313138 - DOI - PMC - PubMed
-
- Zhang D, Jiang X, Fang P, Yan Y, Song J, Gupta S, et al. . Hyperhomocysteinemia Promotes Inflammatory Monocyte Generation and Accelerates Atherosclerosis in Transgenic Cystathionine Beta-Synthase-Deficient Mice. Circulation (2009) 120:1893–902. doi: 10.1161/CIRCULATIONAHA.109.866889 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

