Genomic analysis of Elizabethkingia species from aquatic environments: Evidence for potential clinical transmission
- PMID: 34988536
- PMCID: PMC8703026
- DOI: 10.1016/j.crmicr.2021.100083
Genomic analysis of Elizabethkingia species from aquatic environments: Evidence for potential clinical transmission
Abstract
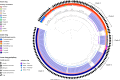
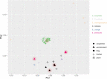
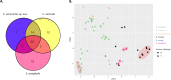
Elizabethkingia species are ubiquitous in aquatic environments, colonize water systems in healthcare settings and are emerging opportunistic pathogens with reports surfacing in 25 countries across six continents. Elizabethkingia infections are challenging to treat, and case fatality rates are high. Chromosomal bla B , bla GOB and bla CME genes encoding carbapenemases and cephalosporinases are unique to Elizabethkingia spp. and reports of concomitant resistance to aminoglycosides, fluoroquinolones and sulfamethoxazole-trimethoprim are known. Here, we characterized whole-genome sequences of 94 Elizabethkingia isolates carrying multiple wide-spectrum metallo-β-lactamase (bla B and bla GOB) and extended-spectrum serine‑β-lactamase (bla CME) genes from Australian aquatic environments and performed comparative phylogenomic analyses against national clinical and international strains. qPCR was performed to quantify the levels of Elizabethkingia species in the source environments. Antibiotic MIC testing revealed significant resistance to carbapenems and cephalosporins but susceptibility to fluoroquinolones, tetracyclines and trimethoprim-sulfamethoxazole. Phylogenetics show that three environmental E. anophelis isolates are closely related to E. anophelis from Australian clinical isolates (∼36 SNPs), and a new species, E. umeracha sp. novel, was discovered. Genomic signatures provide insight into potentially shared origins and a capacity to transfer mobile genetic elements with both national and international isolates.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L.V., Cheng A.A., Liu S., Min S.Y., Miroshnichenko A., Tran H.K., Werfalli R.E., Nasir J.A., Oloni M., Speicher D.J., Florescu A., Singh B., Faltyn M., Hernandez-Koutoucheva A., Sharma A.N., Bordeleau E., Pawlowski A.C., Zubyk H.L., Dooley D., Griffiths E., Maguire F., Winsor G.L., Beiko R.G., Brinkman F.S.L., Hsiao W.W.L., Domselaar G.V., McArthur A.G. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019;935 doi: 10.1093/nar/gkz935. - DOI - PMC - PubMed
-
- Ante V.M., Farris L.C., Saputra E.P., Hall A.J., O'Bier N.S., Oliva Chávez A.S., Marconi R.T., Lybecker M.C., Hyde J.A. The borrelia burgdorferi adenylate cyclase, CyaB, is important for virulence factor production and mammalian infection. Front. Microbiol. 2021;12:1121. doi: 10.3389/fmicb.2021.676192. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

