Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance
- PMID: 34989238
- PMCID: PMC8751645
- DOI: 10.1021/acs.jcim.1c01451
Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance
Abstract
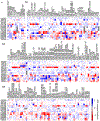
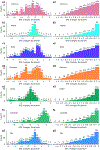
The latest severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant Omicron (B.1.1.529) has ushered panic responses around the world due to its contagious and vaccine escape mutations. The essential infectivity and antibody resistance of the SARS-CoV-2 variant are determined by its mutations on the spike (S) protein receptor-binding domain (RBD). However, a complete experimental evaluation of Omicron might take weeks or even months. Here, we present a comprehensive quantitative analysis of Omicron's infectivity, vaccine breakthrough, and antibody resistance. An artificial intelligence (AI) model, which has been trained with tens of thousands of experimental data and extensively validated by experimental results on SARS-CoV-2, reveals that Omicron may be over 10 times more contagious than the original virus or about 2.8 times as infectious as the Delta variant. On the basis of 185 three-dimensional (3D) structures of antibody-RBD complexes, we unveil that Omicron may have an 88% likelihood to escape current vaccines. The U.S. Food and Drug Administration (FDA)-approved monoclonal antibodies (mAbs) from Eli Lilly may be seriously compromised. Omicron may also diminish the efficacy of mAbs from AstraZeneca, Regeneron mAb cocktail, Celltrion, and Rockefeller University. However, its impacts on GlaxoSmithKline's sotrovimab appear to be mild. Our work calls for new strategies to develop the next generation mutation-proof SARS-CoV-2 vaccines and antibodies.
Figures

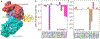
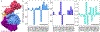
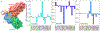
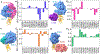
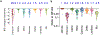

Update of
-
Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance.ArXiv [Preprint]. 2021 Dec 1:arXiv:2112.01318v1. ArXiv. 2021. Update in: J Chem Inf Model. 2022 Jan 24;62(2):412-422. doi: 10.1021/acs.jcim.1c01451. PMID: 34873578 Free PMC article. Updated. Preprint.
References
-
- Li W; Shi Z; Yu M; Ren W; Smith C; Epstein JH; Wang H; Crameri G; Hu Z; Zhang H, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. - PubMed
-
- Qu X-X; Hao P; Song X-J; Jiang S-M; Liu Y-X; Wang P-G; Rao X; Song H-D; Wang S-Y; Zuo Y, et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem 2005, 280, 29588–29595. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

