Evaluating the aerobic xylene-degrading potential of the intrinsic microbial community of a legacy BTEX-contaminated aquifer by enrichment culturing coupled with multi-omics analysis: uncovering the role of Hydrogenophaga strains in xylene degradation
- PMID: 34989990
- PMCID: PMC8993774
- DOI: 10.1007/s11356-021-18300-w
Evaluating the aerobic xylene-degrading potential of the intrinsic microbial community of a legacy BTEX-contaminated aquifer by enrichment culturing coupled with multi-omics analysis: uncovering the role of Hydrogenophaga strains in xylene degradation
Abstract
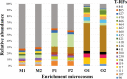
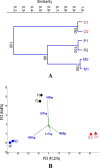
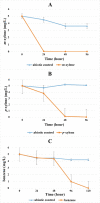
To develop effective bioremediation strategies, it is always important to explore autochthonous microbial community diversity using substrate-specific enrichment. The primary objective of this present study was to reveal the diversity of aerobic xylene-degrading bacteria at a legacy BTEX-contaminated site where xylene is the predominant contaminant, as well as to identify potential indigenous strains that could effectively degrade xylenes, in order to better understand the underlying facts about xylene degradation using a multi-omics approach. Henceforward, parallel aerobic microcosms were set up using different xylene isomers as the sole carbon source to investigate evolved bacterial communities using both culture-dependent and independent methods. Research outcome showed that the autochthonous community of this legacy BTEX-contaminated site has the capability to remove all of the xylene isomers from the environment aerobically employing different bacterial groups for different xylene isomers. Interestingly, polyphasic analysis of the enrichments disclose that the community composition of the o-xylene-degrading enrichment community was utterly distinct from that of the m- and p-xylene-degrading enrichments. Although in each of the enrichments Pseudomonas and Acidovorax were the dominant genera, in the case of o-xylene-degrading enrichment Rhodococcus was the main player. Among the isolates, two Hydogenophaga strains, belonging to the same genomic species, were obtained from p-xylene-degrading enrichment, substantially able to degrade aromatic hydrocarbons including xylene isomers aerobically. Comparative whole-genome analysis of the strains revealed different genomic adaptations to aromatic hydrocarbon degradation, providing an explanation on their different xylene isomer-degrading abilities.
Keywords: Aromatic hydrocarbons; Biodegradation; Catechol 2,3-dioxygenase; Groundwater; Hydrogenohaga; Xylene.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

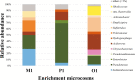

References
-
- Aburto A, Fahy A, Coulon F, et al (2009) Mixed aerobic and anaerobic microbial communities in benzene-contaminated groundwater. J Appl Microbiol 10610.1111/j.1365-2672.2008.04005.x - PubMed
-
- Alrumman SA, Standing DB, Paton GI (2015) Effects of hydrocarbon contamination on soil microbial community and enzyme activity. J King Saud Univ - Sci 2710.1016/j.jksus.2014.10.001
-
- Andreoni V, Gianfreda L (2007) Bioremediation and monitoring of aromatic-polluted habitats. Appl. Microbiol. Biotechnol. 76 - PubMed
MeSH terms
Substances
Grants and funding
- Thematic Excellence Programme 2020/ministry of innovation and technology of hungary
- Institutional Excellence Subprogramme (TKP2020-IKA-12)/ministry of innovation and technology of hungary
- FK 134439/"otka" young researcher excellence programme
- Stipendium Hungaricum Scholarship Programme/tempus public foundation
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

