Mutational analysis of Aedes aegypti Dicer 2 provides insights into the biogenesis of antiviral exogenous small interfering RNAs
- PMID: 34990484
- PMCID: PMC8769306
- DOI: 10.1371/journal.ppat.1010202
Mutational analysis of Aedes aegypti Dicer 2 provides insights into the biogenesis of antiviral exogenous small interfering RNAs
Abstract
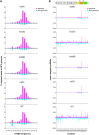
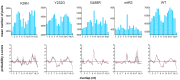
The exogenous small interfering RNA (exo-siRNA) pathway is a key antiviral mechanism in the Aedes aegypti mosquito, a widely distributed vector of human-pathogenic arboviruses. This pathway is induced by virus-derived double-stranded RNAs (dsRNA) that are cleaved by the ribonuclease Dicer 2 (Dcr2) into predominantly 21 nucleotide (nt) virus-derived small interfering RNAs (vsiRNAs). These vsiRNAs are used by the effector protein Argonaute 2 within the RNA-induced silencing complex to cleave target viral RNA. Dcr2 contains several domains crucial for its activities, including helicase and RNase III domains. In Drosophila melanogaster Dcr2, the helicase domain has been associated with binding to dsRNA with blunt-ended termini and a processive siRNA production mechanism, while the platform-PAZ domains bind dsRNA with 3' overhangs and subsequent distributive siRNA production. Here we analyzed the contributions of the helicase and RNase III domains in Ae. aegypti Dcr2 to antiviral activity and to the exo-siRNA pathway. Conserved amino acids in the helicase and RNase III domains were identified to investigate Dcr2 antiviral activity in an Ae. aegypti-derived Dcr2 knockout cell line by reporter assays and infection with mosquito-borne Semliki Forest virus (Togaviridae, Alphavirus). Functionally relevant amino acids were found to be conserved in haplotype Dcr2 sequences from field-derived Ae. aegypti across different continents. The helicase and RNase III domains were critical for silencing activity and 21 nt vsiRNA production, with RNase III domain activity alone determined to be insufficient for antiviral activity. Analysis of 21 nt vsiRNA sequences (produced by functional Dcr2) to assess the distribution and phasing along the viral genome revealed diverse yet highly consistent vsiRNA pools, with predominantly short or long sequence overlaps including 19 nt overlaps (the latter representing most likely true Dcr2 cleavage products). Combined with the importance of the Dcr2 helicase domain, this suggests that the majority of 21 nt vsiRNAs originate by processive cleavage. This study sheds new light on Ae. aegypti Dcr2 functions and properties in this important arbovirus vector species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

