Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization
- PMID: 34990590
- PMCID: PMC10089621
- DOI: 10.1016/j.immuni.2021.12.003
Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization
Abstract
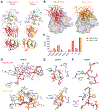
The high genetic diversity of hepatitis C virus (HCV) complicates effective vaccine development. We screened a cohort of 435 HCV-infected individuals and found that 2%-5% demonstrated outstanding HCV-neutralizing activity. From four of these patients, we isolated 310 HCV antibodies, including neutralizing antibodies with exceptional breadth and potency. High neutralizing activity was enabled by the use of the VH1-69 heavy-chain gene segment, somatic mutations within CDRH1, and CDRH2 hydrophobicity. Structural and mutational analyses revealed an important role for mutations replacing the serines at positions 30 and 31, as well as the presence of neutral and hydrophobic residues at the tip of the CDRH3. Based on these characteristics, we computationally created a de novo antibody with a fully synthetic VH1-69 heavy chain that efficiently neutralized multiple HCV genotypes. Our findings provide a deep understanding of the generation of broadly HCV-neutralizing antibodies that can guide the design of effective vaccine candidates.
Keywords: HCV; V(H)1-69; elite neutralizer; hepatitis C virus; machine learning; monoclonal antibody; neutralizing antibody; single B cell analysis; somatic mutation.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Reported antibodies are in the process of being patented.
Figures





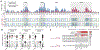
Comment in
-
HCV neutralization goes elite.Immunity. 2022 Feb 8;55(2):195-197. doi: 10.1016/j.immuni.2022.01.010. Immunity. 2022. PMID: 35139349
-
Convergent evolution and targeting of diverse E2 epitopes by human broadly neutralizing antibodies are associated with HCV clearance.Immunity. 2024 Apr 9;57(4):890-903.e6. doi: 10.1016/j.immuni.2024.03.001. Epub 2024 Mar 21. Immunity. 2024. PMID: 38518779 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

