The renal clear cell carcinoma immune landscape
- PMID: 34991061
- PMCID: PMC8740459
- DOI: 10.1016/j.neo.2021.12.007
The renal clear cell carcinoma immune landscape
Abstract
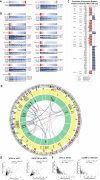
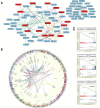
A comprehensive evaluation of the clear cell renal cell carcinoma (ccRCC) immune landscape was found using 584 RNA-sequencing datasets from The Cancer Genome Atlas (TCGA), we identified 17 key dysregulated immune-associated genes in ccRCC based on association with clinical variables and important immune pathways. Of the numerous findings from our analyses, we found that several of the 17 key dysregulated genes are heavily involved in interleukin and NF-kB signaling and that somatic copy number alteration (SCNA) hotspots may be causally associated with gene dysregulation. More importantly, we also found that key immune-associated genes and pathways are strongly upregulated in ccRCC. Our study may lend novel insights into the clinical implications of immune dysregulation in ccRCC and suggests potential immunotherapeutic targets for further evaluation.
Keywords: Renal clear cell carcinoma; TCGA.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures