Genomic predictors of response to PD-1 inhibition in children with germline DNA replication repair deficiency
- PMID: 34992263
- PMCID: PMC8799468
- DOI: 10.1038/s41591-021-01581-6
Genomic predictors of response to PD-1 inhibition in children with germline DNA replication repair deficiency
Abstract
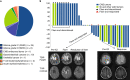
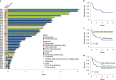
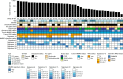
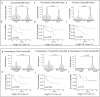
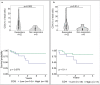
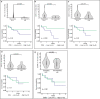
Cancers arising from germline DNA mismatch repair deficiency or polymerase proofreading deficiency (MMRD and PPD) in children harbour the highest mutational and microsatellite insertion-deletion (MS-indel) burden in humans. MMRD and PPD cancers are commonly lethal due to the inherent resistance to chemo-irradiation. Although immune checkpoint inhibitors (ICIs) have failed to benefit children in previous studies, we hypothesized that hypermutation caused by MMRD and PPD will improve outcomes following ICI treatment in these patients. Using an international consortium registry study, we report on the ICI treatment of 45 progressive or recurrent tumors from 38 patients. Durable objective responses were observed in most patients, culminating in a 3 year survival of 41.4%. High mutation burden predicted response for ultra-hypermutant cancers (>100 mutations per Mb) enriched for combined MMRD + PPD, while MS-indels predicted response in MMRD tumors with lower mutation burden (10-100 mutations per Mb). Furthermore, both mechanisms were associated with increased immune infiltration even in 'immunologically cold' tumors such as gliomas, contributing to the favorable response. Pseudo-progression (flare) was common and was associated with immune activation in the tumor microenvironment and systemically. Furthermore, patients with flare who continued ICI treatment achieved durable responses. This study demonstrates improved survival for patients with tumors not previously known to respond to ICI treatment, including central nervous system and synchronous cancers, and identifies the dual roles of mutation burden and MS-indels in predicting sustained response to immunotherapy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

