Imaging-Based Machine Learning Analysis of Patient-Derived Tumor Organoid Drug Response
- PMID: 34993134
- PMCID: PMC8724556
- DOI: 10.3389/fonc.2021.771173
Imaging-Based Machine Learning Analysis of Patient-Derived Tumor Organoid Drug Response
Abstract
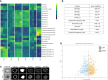
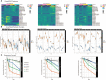
Three-quarters of compounds that enter clinical trials fail to make it to market due to safety or efficacy concerns. This statistic strongly suggests a need for better screening methods that result in improved translatability of compounds during the preclinical testing period. Patient-derived organoids have been touted as a promising 3D preclinical model system to impact the drug discovery pipeline, particularly in oncology. However, assessing drug efficacy in such models poses its own set of challenges, and traditional cell viability readouts fail to leverage some of the advantages that the organoid systems provide. Consequently, phenotypically evaluating complex 3D cell culture models remains difficult due to intra- and inter-patient organoid size differences, cellular heterogeneities, and temporal response dynamics. Here, we present an image-based high-content assay that provides object level information on 3D patient-derived tumor organoids without the need for vital dyes. Leveraging computer vision, we segment and define organoids as independent regions of interest and obtain morphometric and textural information per organoid. By acquiring brightfield images at different timepoints in a robust, non-destructive manner, we can track the dynamic response of individual organoids to various drugs. Furthermore, to simplify the analysis of the resulting large, complex data files, we developed a web-based data visualization tool, the Organoizer, that is available for public use. Our work demonstrates the feasibility and utility of using imaging, computer vision and machine learning to determine the vital status of individual patient-derived organoids without relying upon vital dyes, thus taking advantage of the characteristics offered by this preclinical model system.
Keywords: drug response; high content imaging; label-free analysis; machine learning; patient-derived organoids (PDO).
Copyright © 2021 Spiller, Ung, Kim, Patsch, Lau, Strelez, Doshi, Choung, Choi, Juarez Rosales, Lenz, Matasci and Mumenthaler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

