Patient derived models of bladder cancer enrich the signal of the tumor cell transcriptome facilitating the analysis of the tumor cell compartment
- PMID: 34993263
- PMCID: PMC8727788
Patient derived models of bladder cancer enrich the signal of the tumor cell transcriptome facilitating the analysis of the tumor cell compartment
Abstract
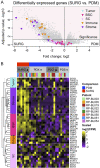
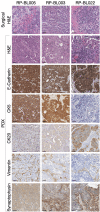
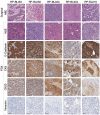
The evolving paradigm of the molecular classification of bladder cancer requires models that represent the classifications with less heterogeneity. Robust transcriptome based molecular classifications are essential to address tumor heterogeneity. Patient derived models (PDMs) are a powerful preclinical tool to study specific tumor compartments. We tested if the consensus molecular subtype analysis was applicable to PDMs and evaluated the tumor compartment each model represents. PDMs derived from surgical specimens were established as xenografts (PDX), organoids (PDO), and spheroids (PDS). The surgical specimens and PDMs were molecularly characterized by RNA sequencing. PDMs that were established in immune deficient mice or in vitro significantly downregulated transcripts related to the immune and stromal compartments compared to the surgical specimens. However, PDMs upregulate a patient-specific bladder cancer cell signal which allowed for analysis of cancer cell pathways independent of the tumor microenvironment. Based on transcriptomic signatures, PDMs are more similar to their surgical specimen than the model type; indicating that the PDMs retained unique features of the tumor from which the PDM was derived. When comparing models, PDX models were the most similar to the surgical specimen, while PDO and PDS models were most similar to each other. When the consensus molecular subtype classification system was applied to both the surgical samples and the three PDMs, good concordance was found between all samples indicating that this system of classification can be applied to PDO and PDS models. PDMs reduce tumor heterogeneity and allow analysis of tumor cells while maintaining the gene expression profile representative of the original tumor.
Keywords: Bladder cancer; epithelial to mesenchymal transition (EMT); organoid; patient derived models; spheroid; xenograft.
AJCEU Copyright © 2021.
Conflict of interest statement
None.
Figures



References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. - PubMed
-
- Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, Castro MAA, Gibb EA, Kanchi RS, Gordenin DA, Shukla SA, Sanchez-Vega F, Hansel DE, Czerniak BA, Reuter VE, Su X, de Sa Carvalho B, Chagas VS, Mungall KL, Sadeghi S, Pedamallu CS, Lu Y, Klimczak LJ, Zhang J, Choo C, Ojesina AI, Bullman S, Leraas KM, Lichtenberg TM, Wu CJ, Schultz N, Getz G, Meyerson M, Mills GB, McConkey DJ TCGA Research Network. Weinstein JN, Kwiatkowski DJ, Lerner SP. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–556. e525. - PMC - PubMed
-
- Skowron KB, Pitroda SP, Namm JP, Balogun O, Beckett MA, Zenner ML, Fayanju O, Huang X, Fernandez C, Zheng W, Qiao G, Chin R, Kron SJ, Khodarev NN, Posner MC, Steinberg GD, Weichselbaum RR. Basal tumor cell isolation and patient-derived xenograft engraftment identify high-risk clinical bladder cancers. Sci Rep. 2016;6:35854. - PMC - PubMed
-
- Wei L, Chintala S, Ciamporcero E, Ramakrishnan S, Elbanna M, Wang J, Hu Q, Glenn ST, Murakami M, Liu L, Gomez EC, Sun Y, Conroy J, Miles KM, Malathi K, Ramaiah S, Anbarasu A, Woloszynska-Read A, Johnson CS, Conroy J, Liu S, Morrison CD, Pili R. Genomic profiling is predictive of response to cisplatin treatment but not to PI3K inhibition in bladder cancer patient-derived xenografts. Oncotarget. 2016;7:76374–76389. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
