mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant
- PMID: 34995482
- PMCID: PMC8733787
- DOI: 10.1016/j.cell.2021.12.033
mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant
Abstract
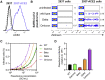
Recent surveillance has revealed the emergence of the SARS-CoV-2 Omicron variant (BA.1/B.1.1.529) harboring up to 36 mutations in spike protein, the target of neutralizing antibodies. Given its potential to escape vaccine-induced humoral immunity, we measured the neutralization potency of sera from 88 mRNA-1273, 111 BNT162b, and 40 Ad26.COV2.S vaccine recipients against wild-type, Delta, and Omicron SARS-CoV-2 pseudoviruses. We included individuals that received their primary series recently (<3 months), distantly (6-12 months), or an additional "booster" dose, while accounting for prior SARS-CoV-2 infection. Remarkably, neutralization of Omicron was undetectable in most vaccinees. However, individuals boosted with mRNA vaccines exhibited potent neutralization of Omicron, only 4-6-fold lower than wild type, suggesting enhanced cross-reactivity of neutralizing antibody responses. In addition, we find that Omicron pseudovirus infects more efficiently than other variants tested. Overall, this study highlights the importance of additional mRNA doses to broaden neutralizing antibody responses against highly divergent SARS-CoV-2 variants.
Keywords: COVID-19; Delta; Omicron; SARS-CoV-2; breadth; infectivity; neutralizing antibodies; spike; vaccination; variants.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Update of
-
mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant.medRxiv [Preprint]. 2021 Dec 14:2021.12.14.21267755. doi: 10.1101/2021.12.14.21267755. medRxiv. 2021. Update in: Cell. 2022 Feb 3;185(3):457-466.e4. doi: 10.1016/j.cell.2021.12.033. PMID: 34931201 Free PMC article. Updated. Preprint.
Comment in
-
Omicron, the great escape artist.Nat Rev Immunol. 2022 Feb;22(2):75. doi: 10.1038/s41577-022-00676-6. Nat Rev Immunol. 2022. PMID: 35017722 Free PMC article.
-
Omicron's message on vaccines: Boosting begets breadth.Cell. 2022 Feb 3;185(3):411-413. doi: 10.1016/j.cell.2022.01.006. Epub 2022 Jan 14. Cell. 2022. PMID: 35065712 Free PMC article.
-
Beyond gut instincts: Microbe survival depends on sugars and butyrate.Cell. 2022 Feb 3;185(3):414-416. doi: 10.1016/j.cell.2021.12.037. Cell. 2022. PMID: 35120661
-
Vaccination versus SARS-CoV-2 Omicron: three vaccine doses win the battle.Signal Transduct Target Ther. 2022 Apr 27;7(1):140. doi: 10.1038/s41392-022-01000-3. Signal Transduct Target Ther. 2022. PMID: 35477929 Free PMC article. No abstract available.
References
-
- Bajema K.L., Dahl R.M., Evener S.L., Prill M.M., Rodriguez-Barradas M.C., Marconi V.C., Beenhouwer D.O., Holodniy M., Lucero-Obusan C., Brown S.T., et al. Comparative effectiveness and antibody responses to moderna and Pfizer-BioNTech COVID-19 vaccines among hospitalized veterans—five Veterans Affairs Medical Centers, United States, February 1–September 30, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1700–1705. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

