Increased circulating butyrate and ursodeoxycholate during probiotic intervention in humans with type 2 diabetes
- PMID: 34996347
- PMCID: PMC8742391
- DOI: 10.1186/s12866-021-02415-8
Increased circulating butyrate and ursodeoxycholate during probiotic intervention in humans with type 2 diabetes
Abstract
Background: An increasing body of evidence implicates the resident gut microbiota as playing a critical role in type 2 diabetes (T2D) pathogenesis. We previously reported significant improvement in postprandial glucose control in human participants with T2D following 12-week administration of a 5-strain novel probiotic formulation ('WBF-011') in a double-blind, randomized, placebo controlled setting (NCT03893422). While the clinical endpoints were encouraging, additional exploratory measurements were needed in order to link the motivating mechanistic hypothesis - increased short-chain fatty acids - with markers of disease.
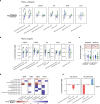
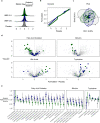
Results: Here we report targeted and untargeted metabolomic measurements on fasting plasma (n = 104) collected at baseline and end of intervention. Butyrate and ursodeoxycholate increased among participants randomized to WBF-011, along with compelling trends between butyrate and glycated haemoglobin (HbA1c). In vitro monoculture experiments demonstrated that the formulation's C. butyricum strain efficiently synthesizes ursodeoxycholate from the primary bile acid chenodeoxycholate during butyrogenic growth. Untargeted metabolomics also revealed coordinated decreases in intermediates of fatty acid oxidation and bilirubin, potential secondary signatures for metabolic improvement. Finally, improvement in HbA1c was limited almost entirely to participants not using sulfonylurea drugs. We show that these drugs can inhibit growth of formulation strains in vitro.
Conclusion: To our knowledge, this is the first description of an increase in circulating butyrate or ursodeoxycholate following a probiotic intervention in humans with T2D, adding support for the possibility of a targeted microbiome-based approach to assist in the management of T2D. The efficient synthesis of UDCA by C. butyricum is also likely of interest to investigators of its use as a probiotic in other disease settings. The potential for inhibitory interaction between sulfonylurea drugs and gut microbiota should be considered carefully in the design of future studies.
Keywords: Akkermansia muciniphila; Anaerobutyricum hallii; Bile acids; Butyrate; Clostridium butyricum; Metabolomics; Short-chain fatty acids; Sulfonylurea; Type 2 diabetes; Ursodeoxycholate.
© 2022. The Author(s).
Conflict of interest statement
All authors are employees and stock/stock option shareholders of Pendulum Therapeutics, Inc. (formerly known as ‘Whole Biome Inc.’). O.K. also owns stock in GlySens, Inc., has stock options in ViaCyte, Inc. and NanoPrecision Medical.
Figures



References
-
- CDC. National Diabetes Statistics Report, 2020: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention (CDC); 2020:2–13. https://www.cdc.gov/diabetes/library/features/diabetes-stat-report.html. Accessed 26 Apr 2021
-
- Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, Everard A. Microbial regulation of organismal energy homeostasis. Nat Metab. 2019;1:34–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

